13.3 Defining the Entire Toolkit
The Hox genes are perhaps the best-known members of the toolkit, but they are just a small family in a much larger group of genes required for the development of the proper numbers, shapes, sizes, and kinds of body parts. Little was known about the rest of the toolkit until the late 1970s and early 1980s, when Christine Nüsslein-Volhard and Eric Wieschaus, working at the Max Planck Institute in Tübingen, Germany, set out to find the genes required for the formation of the segmental organization of the Drosophila embryo and larva.
Until their efforts, most work on fly development focused on viable adult phenotypes and not the embryo. Nüsslein-Volhard and Wieschaus realized that the sorts of genes that they were looking for were probably lethal to embryos or larvae in homozygous mutants. So, they came up with a scheme to search for genes that were required in the zygote (the product of fertilization; Figure 13-12, bottom). They also developed screens to identify those genes with products that function in the egg, before the zygotic genome is active, and that are required for the proper patterning of the embryo. Genes with products provided by the female to the egg are called maternal-effect genes. Mutant phenotypes of strict maternal-effect genes depend only on the genotype of the female (Figure 13-12, top).
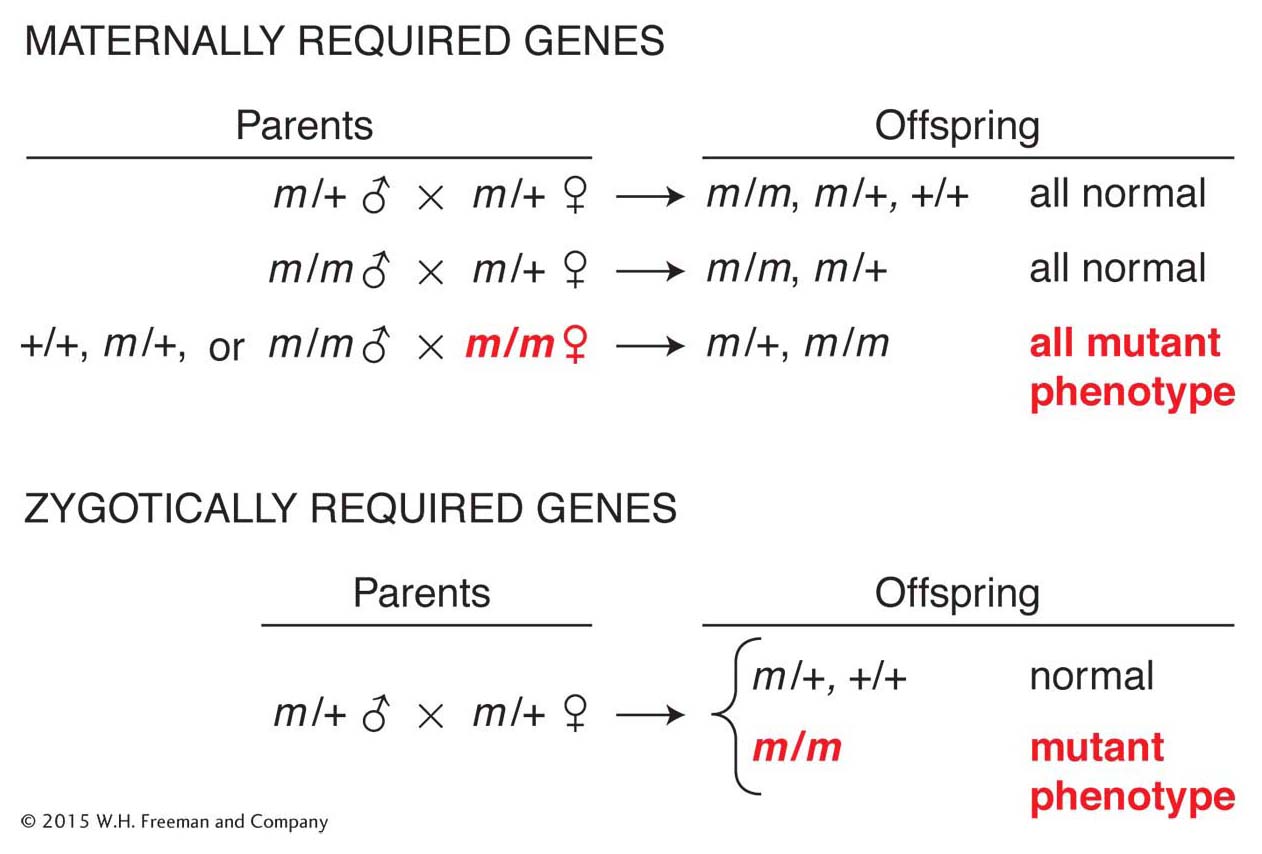
Figure 13-12: Genetic screens for maternally and zygotically required toolkit genes
Figure 13-12: Genetic screens identify whether a gene product functions in the egg or in the zygote. The phenotypes of offspring depend on either (top) the maternal genotype for maternal-effect genes or (bottom) the offspring (zygotic) genotype for zygotically required genes (m, mutant; +, wild type).
In these screens, genes were identified that were necessary to make the proper number and pattern of larval segments, to make its three tissue layers (ectoderm, mesoderm, and endoderm), and to pattern the fine details of an animal’s anatomy. The power of the genetic screens was their systematic nature. By saturating each of a fly’s chromosomes (except the small fourth chromosome) with chemically induced mutations, the researchers were able to identify most genes that were required for the building of the fly. For their pioneering efforts, Nüsslein-Volhard, Wieschaus, and Lewis shared the 1995 Nobel Prize in Physiology or Medicine.
The most striking and telling features of the newly identified mutants were that they showed dramatic but discrete defects in embryo organization or patterning. That is, the dead larva was not an amorphous carcass but exhibited specific, often striking patterning defects. The Drosophila larval body has various features whose number, position, or pattern can serve as landmarks to diagnose or classify the abnormalities in mutant animals. Each locus could thus be classified according to the body axis that it affected and the pattern of defects caused by mutations. Genetic crosses reveal whether the locus is active in the maternal egg or the zygote. Each class of genes appeared to represent different steps in the progressive refinement of the embryonic body plan—from those that affect large regions of the embryo to those with more limited realms of influence.
For any toolkit gene, three pieces of information are key toward understanding gene function: (1) the mutant phenotype, (2) the pattern of gene expression, and (3) the nature of the gene product. Extensive study of a few dozen genes has led to a fairly detailed picture of how each body axis is established and subdivided into segments or germ layers.
The anteroposterior and dorsoventral axes
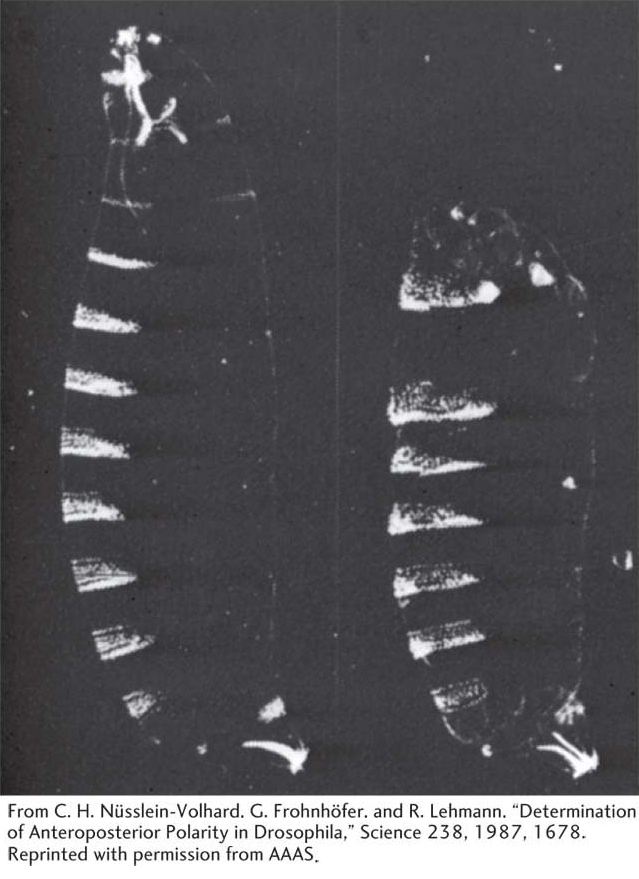
Figure 13-13: Bicoid mutants are missing the anterior region
Figure 13-13: The Bicoid (bcd) maternal-effect gene affects the anterior part of the developing larva. These photomicrographs are of Drosophila larvae that have been prepared to show their hard exoskeletons. Dense structures, such as the segmental denticle bands, appear white. (Left) A normal larva. (Right) A larva from a homozygous bcd mutant female. Head and anterior thoracic structures are missing.
[From C. H. Nüsslein-Volhard, G. Frohnhöfer, and R. Lehmann, “Determination of Anteroposterior Polarity in Drosophila,” Science 238, 1987, 1678. Reprinted with permission from AAAS.]
A few dozen genes are required for proper organization of the anteroposterior body axis of the fly embryo. The genes are grouped into five classes on the basis of their realm of influence on embryonic pattern.
The first class sets up the anteroposterior axis and consists of the maternal-effect genes. A key member of this class is the Bicoid gene. Embryos from Bicoid mutant mothers are missing the anterior region of the embryo (Figure 13-13), telling us that the gene is required for the development of that region.
The next three classes are zygotically active genes required for the development of the segments of the embryo.
The second class contains the gap genes. Each of these genes affects the formation of a contiguous block of segments; mutations in gap genes lead to large gaps in segmentation (Figure 13-14, left).
The third class comprises the pair-rule genes, which act at a double-segment periodicity. Pair-rule mutants are missing part of each pair of segments, but different pair-rule genes affect different parts of each double segment. For example, the even-skipped gene affects one set of segmental boundaries, and the odd-skipped gene affects the complementary set of boundaries (Figure 13-14, middle).
The fourth class consists of the segment-polarity genes, which affect patterning within each segment. Mutants of this class display defects in segment polarity and number (Figure 13-14, right).
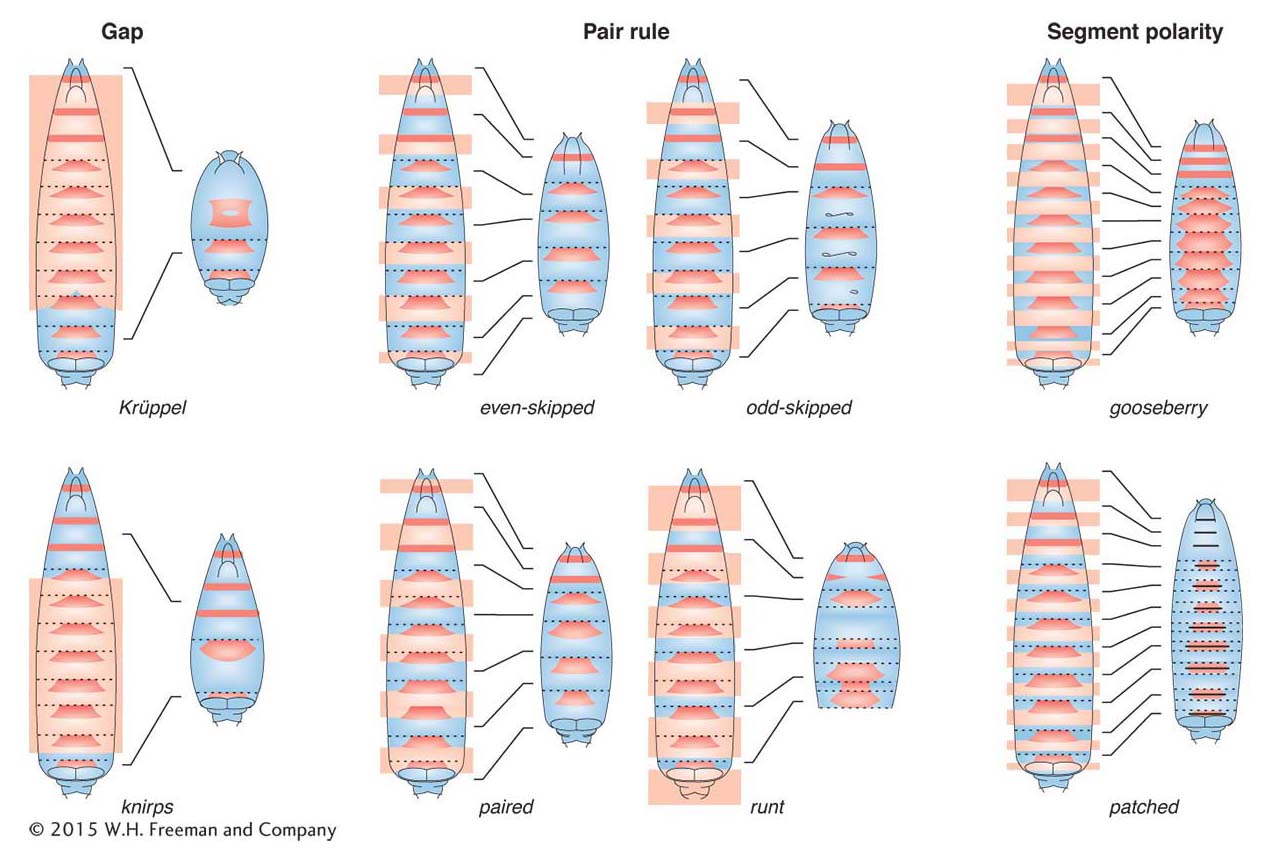
Figure 13-14: Segmentation-gene mutants are missing parts of segments
Figure 13-14: Classes of Drosophila segmentation-gene mutants. These diagrams depict representative gap, pair-rule, and segment-polarity mutants. The red trapezoids are the dense bands of exoskeleton seen in Figure 13-13. The boundary of each segment is indicated by a dotted line. The left-hand diagram of each pair depicts a wild-type larva, and the right-hand diagram depicts the pattern formed in a given mutant. The shaded light orange regions on the wild-type diagrams indicate the domains of the larva that are missing or affected in the mutant.
The fifth class of genes determines the fate of each segment.
The fifth class includes the Hox genes already discussed; Hox mutants do not affect segment number, but they alter the appearance of one or more segments.
Expression of toolkit genes
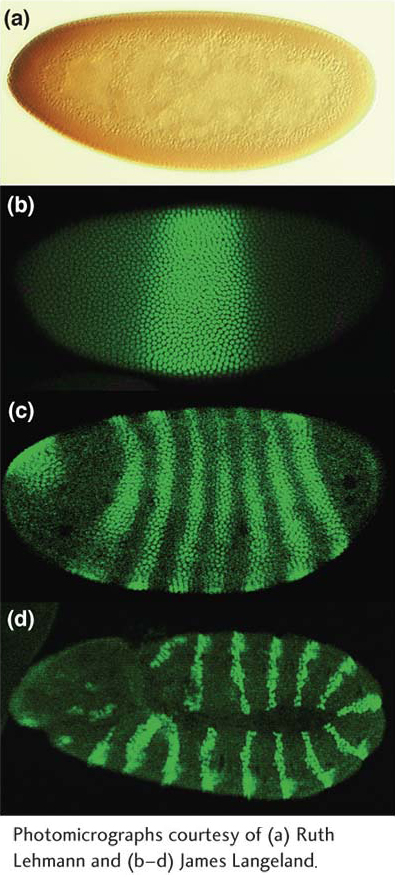
Figure 13-15: Expression of anteroposterior-axis-patterning proteins
Figure 13-15: Patterns of toolkit-gene expression correspond to mutant phenotypes. Drosophila embryos have been stained with antibodies to the (a) maternally derived Bicoid protein, (b) Krüppel gap protein, (c) Hairy pair-rule protein, and (d) Engrailed segment-polarity protein and visualized by immunoenzymatic (staining is brown) (a) or immunofluorescence (staining is green) (b–d) methods. Each protein is localized to nuclei in regions of the embryo that are affected by mutations in the respective genes.
[Photomicrographs courtesy of (a) Ruth Lehmann and (b–d) James Langeland.]
To understand the relation between genes and mutant phenotype, we must know the timing and location of gene-expression patterns and the molecular nature of the gene products. The patterns of expression of the toolkit genes turn out to vividly correspond to their phenotypes, inasmuch as they are often precisely correlated with the parts of the developing body that are altered in mutants. Each gene is expressed in a region that can be mapped to specific coordinates along either axis of the embryo. For example, the maternal-effect Bicoid protein is expressed in a graded pattern emanating from the anterior pole of the early embryo, the section of the embryo missing in mutants (Figure 13-15a). Similarly, the gap proteins are expressed in blocks of cells that correspond to the future positions of the segments that are missing in respective gap-gene mutants (Figure 13-15b). The pair-rule proteins are expressed in striking striped patterns: one transverse stripe is expressed per every 2 segments, in a total of 7 stripes covering the 14 future body segments (the position and periodicity of the stripes correspond to the periodicity of defects in mutant larvae), as shown in Figure 13-15c. Many segment-polarity genes are expressed in stripes of cells within each segment, 14 stripes in all corresponding to 14 body segments (Figure 13-15d). Note that the domains of gene expression become progressively more refined as development proceeds: genes are expressed first in large regions (gap proteins), then in stripes from three to four cells wide (pair-rule proteins), and then in stripes from one to two cells wide (segment-polarity proteins).
In addition to what we have learned from the spatial patterns of toolkit-gene expression, the order of toolkit-gene expression over time is logical. The maternal-effect Bicoid protein appears before the zygotic gap proteins, which are expressed before the 7-striped patterns of pair-rule proteins appear, which in turn precede the 14-striped patterns of segment-polarity proteins. The order of gene expression and the progressive refinement of domains within the embryo reveal that the making of the body plan is a step-by-step process, with major subdivisions of the body outlined first and then refined until a fine-grain pattern is established. The order of gene action further suggests that the expression of one set of genes might govern the expression of the succeeding set of genes.
One clue that this progression is indeed the case comes from analyzing the effects of mutations in toolkit genes on the expression of other toolkit genes. For example, in embryos from Bicoid mutant mothers, the expression of several gap genes is altered, as well as that of pair-rule and segment-polarity genes. This finding suggests that the Bicoid protein somehow (directly or indirectly) influences the regulation of gap genes.
Another clue that the expression of one set of genes might govern the expression of the succeeding set of genes comes from examining the protein products. Inspection of the Bicoid protein sequence reveals that it contains a homeodomain, related to but distinct from those of Hox proteins. Thus, Bicoid has the properties of a DNA-binding transcription factor. Each gap gene also encodes a transcription factor, as does each pair-rule gene, several segment-polarity genes, and, as described earlier, all Hox genes. These transcription factors include representatives of most known families of sequence-specific DNA-binding proteins; so, although there is no restriction concerning to which family they may belong, many early-acting toolkit proteins are transcription factors. Those that are not transcription factors tend to be components of signaling pathways (Table 13-1). These pathways, shown in generic form in Figure 13-16, mediate ligand-induced signaling processes between cells, and their output generally leads to gene activation or repression. Thus, most toolkit proteins either directly (as transcription factors) or indirectly (as components of signaling pathways) affect gene regulation.
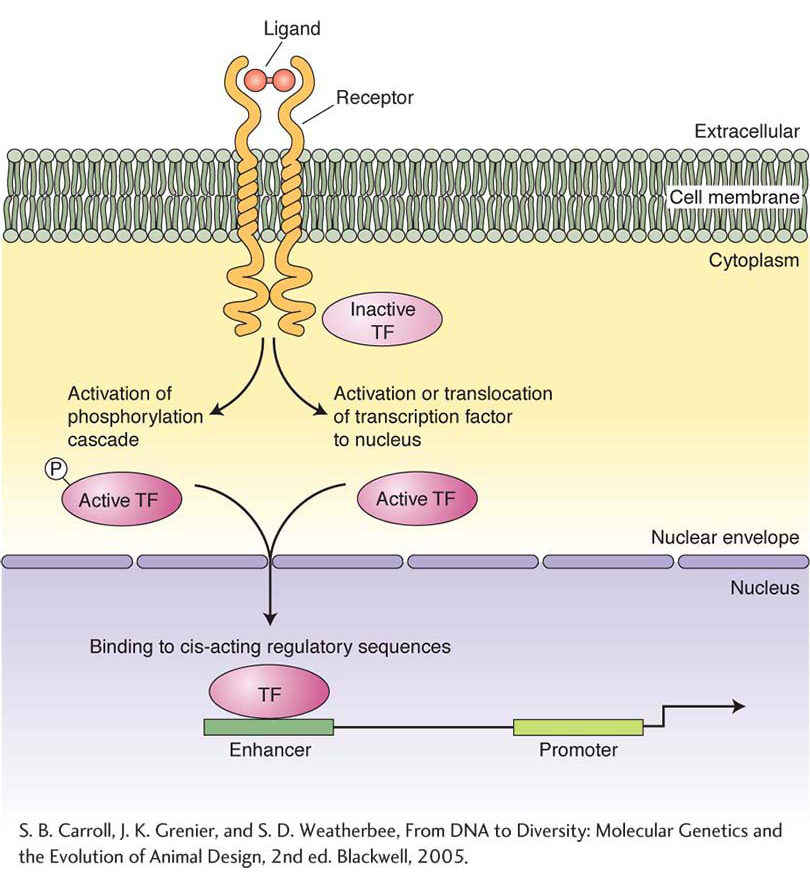
Figure 13-16: A typical signal-transduction pathway
Figure 13-16: Most signaling pathways operate through similar logic but have different protein components and signal-transduction mechanisms. Signaling begins when a ligand binds to a membrane-bound receptor, leading to the release or activation of intracellular proteins. Receptor activation often leads to the modification of inactive transcription factors (TF). The modified transcription factors are translocated to the cell nucleus, where they bind to cis-acting regulatory DNA sequences or to DNA-binding proteins and regulate the level of target-gene transcription.
[S. B. Carroll, J. K. Grenier, and S. D. Weatherbee, From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design, 2nd ed. Blackwell, 2005.]
|
|
|
|
Role(s) in early development |
|
|
|
Transcription factor—zinc-finger protein |
|
|
|
|
Transcription factor—zinc-finger protein |
|
|
|
|
Transcription factor—steroid receptor-type protein |
|
|
|
|
Transcription factor—homeodomain protein |
|
|
|
|
Transcription factor—homeodomain protein |
|
|
|
|
Transcription factor—zinc-finger protein |
|
|
|
|
Transcription factor—PHOX protein |
|
|
|
|
Transcription factor—homeodomain protein |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Transcription factor—homeodomain protein |
|
|
|
|
Transcription factor—homeodomain protein |
|
|
|
|
Transcription factor—homeodomain protein |
|
|
|
|
Transcription factor—homeodomain protein |
|
Table 13-1: Examples of Drosophila A–P Axis Genes That Contribute to Pattern Formation
KEY CONCEPT
Most toolkit proteins are transcription factors or components of ligand-mediated signal-transduction pathways.The same principles apply to the making of the dorsoventral body axis as apply to the anteroposterior axis. The dorsoventral axis also is subdivided into regions. Several maternal-effect genes, such as dorsal, are required to establish these regions at distinct positions from the dorsal (top) to ventral (bottom) side of the embryo. Dorsal mutants are “dorsalized” and lack ventral structures (such as the mesoderm and nervous system). A handful of genes that are activated in the zygote are also required for the subdivision of the dorsoventral axis.
The product of the dorsal maternal-effect gene is a transcription factor—the Dorsal protein. This protein is expressed in a gradient along the dorsoventral axis, with its highest level of accumulation in ventral cells (Figure 13-17a). The gradient establishes subregions of differing Dorsal concentration. In each subregion, a different set of zygotic genes are expressed that contribute to dorsoventral patterning. The sets of zygotic genes expressed define regions that give rise to particular tissue layers, such as the mesoderm and neuroectoderm (the part of the ectoderm that gives rise to the ventral nervous system), as shown in Figure 13-17b.
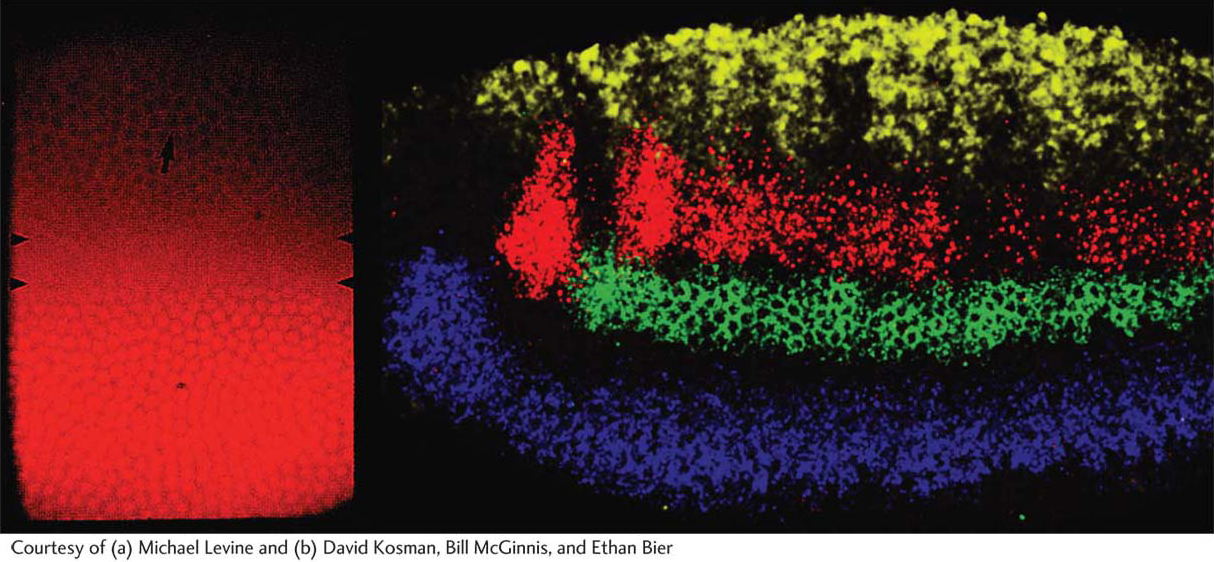
Figure 13-17: Expression of dorsoventral-axis-patterning genes
Figure 13-17: Expression domains of specific dorsoventral-axis-patterning genes correspond to particular future tissue layers. (a) The maternally derived Dorsal protein is expressed in a gradient, with the highest concentration of Dorsal in the nuclei of ventral cells (bottom of photograph). (b) The expression of four zygotic dorsoventral-axis-patterning genes revealed by in situ hybridization to RNA. In this lateral view, the domains of the decapentaplegic (yellow), muscle segment homeobox (red), intermediate neuroblasts defective (green), and ventral neuroblasts defective (blue) genes are revealed.
[Courtesy of (a) Michael Levine and (b) David Kosman, Bill McGinnis, and Ethan Bier.]
The genetic control of development, then, is fundamentally a matter of gene regulation in space and over time. How does the turning on and off of toolkit genes build animal form? And how is it choreographed during development? To answer these questions, we will examine the interactions among fly toolkit proteins and genes in more detail. The mechanisms that we will see for controlling toolkit-gene expression in the Drosophila embryo have emerged as models for the spatial regulation of gene expression in animal development in general.





