17.1 Changes in Chromosome Number
In genetics as a whole, few topics impinge on human affairs quite so directly as that of changes in the number of chromosomes present in our cells. Foremost is the fact that a group of common genetic disorders results from the presence of an abnormal number of chromosomes. Although this group of disorders is small, it accounts for a large proportion of the genetically determined health problems that afflict humans. Also of relevance to humans is the role of chromosome mutations in plant breeding: plant breeders have routinely manipulated chromosome number to improve commercially important agricultural crops.
619
Changes in chromosome number are of two basic types: changes in whole chromosome sets, resulting in a condition called aberrant euploidy, and changes in parts of chromosome sets, resulting in a condition called aneuploidy.
Aberrant euploidy
Organisms with multiples of the basic chromosome set (genome) are referred to as euploid. You learned in earlier chapters that familiar eukaryotes such as plants, animals, and fungi carry in their cells either one chromosome set (haploidy) or two chromosome sets (diploidy). In these species, both the haploid and the diploid states are cases of normal euploidy. Organisms that have more or fewer than the normal number of sets are aberrant euploids. Polyploids are individual organisms that have more than two chromosome sets. They can be represented by 3n (triploid), 4n (tetraploid), 5n (pentaploid), 6n (hexaploid), and so forth. (The number of chromosome sets is called the ploidy or ploidy level.) An individual member of a normally diploid species that has only one chromosome set (n) is called a monoploid to distinguish it from an individual member of a normally haploid species (also n). Examples of these conditions are shown in the first four rows of Table 17-1.
|
Name |
Designation |
Constitution |
Number of chromosomes |
|---|---|---|---|
|
Euploids |
|||
|
Monoploid |
n |
A B C |
3 |
|
Diploid |
2n |
AA BB CC |
6 |
|
Triploid |
3n |
AAA BBB CCC |
9 |
|
Tetraploid |
4n |
AAAA BBBB CCCC |
12 |
|
Aneuploids |
|||
|
Monosomic |
2n − 1 |
A BB CC |
5 |
|
AA B CC |
5 |
||
|
AA BB C |
5 |
||
|
Trisomic |
2n + 1 |
AAA BB CC |
7 |
|
AA BBB CC |
7 |
||
|
AA BB CCC |
7 |
||
Monoploids Male bees, wasps, and ants are monoploid. In the normal life cycles of these insects, males develop by parthenogenesis (the development of a specialized type of unfertilized egg into an embryo without the need for fertilization). In most other species, however, monoploid zygotes fail to develop. The reason is that virtually all members of a diploid species carry a number of deleterious recessive mutations, together called a genetic load. The deleterious recessive alleles are masked by wild-
620
Polyploids Polyploidy is very common in plants but rarer in animals (for reasons that we will consider later). Indeed, an increase in the number of chromosome sets has been an important factor in the origin of new plant species. The evidence for this benefit is that above a haploid number of about 12, even numbers of chromosomes are much more common than odd numbers. This pattern is a consequence of the polyploid origin of many plant species, because doubling and redoubling of a number can give rise only to even numbers. Animal species do not show such a distribution, owing to the relative rareness of polyploid animals.
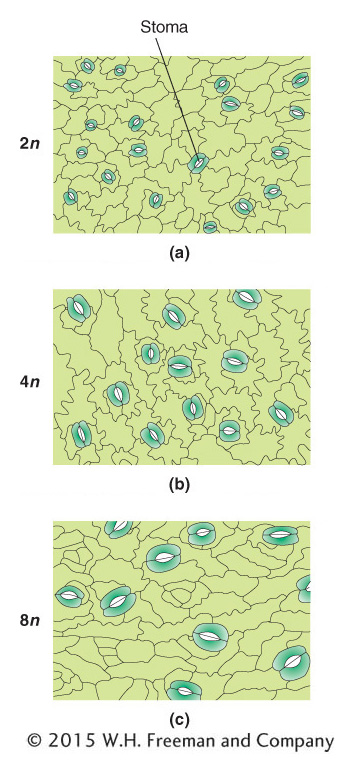
In aberrant euploids, there is often a correlation between the number of copies of the chromosome set and the size of the organism. A tetraploid organism, for example, typically looks very similar to its diploid counterpart in its proportions, except that the tetraploid is bigger, both as a whole and in its component parts. The higher the ploidy level, the larger the size of the organism (Figure 17-3).
KEY CONCEPT
Polyploids are often larger and have larger component parts than their diploid relatives.In the realm of polyploids, we must distinguish between autopolyploids, which have multiple chromosome sets originating from within one species, and allopolyploids, which have sets from two or more different species. Allopolyploids form only between closely related species; however, the different chromosome sets are only homeologous (partly homologous), not fully homologous as they are in autopolyploids.
Autopolyploids Triploids (3n) are usually autopolyploids. They arise spontaneously in nature, but they can be constructed by geneticists from the cross of a 4n (tetraploid) and a 2n (diploid). The 2n and the n gametes produced by the tetraploid and the diploid, respectively, unite to form a 3n triploid. Triploids are characteristically sterile. The problem (which is also true of monoploids) lies in the presence of unpaired chromosomes at meiosis. The molecular mechanisms for synapsis, or true pairing, dictate that, in a triploid, pairing can take place between only two of the three chromosomes of each type (Figure 17-4). Paired homologs (bivalents) segregate to opposite poles, but the unpaired homologs (univalents) pass to either pole randomly. In a trivalent, a paired group of three, the paired centromeres segregate as a bivalent and the unpaired one as a univalent. These segregations take place for every chromosome threesome; so, for any chromosomal type, the gamete could receive either one or two chromosomes. It is unlikely that a gamete will receive two for every chromosomal type or that it will receive one for every chromosomal type. Hence, the likelihood is that gametes will have chromosome numbers intermediate between the haploid and the diploid number; such genomes are of a type called aneuploid (“not euploid”).
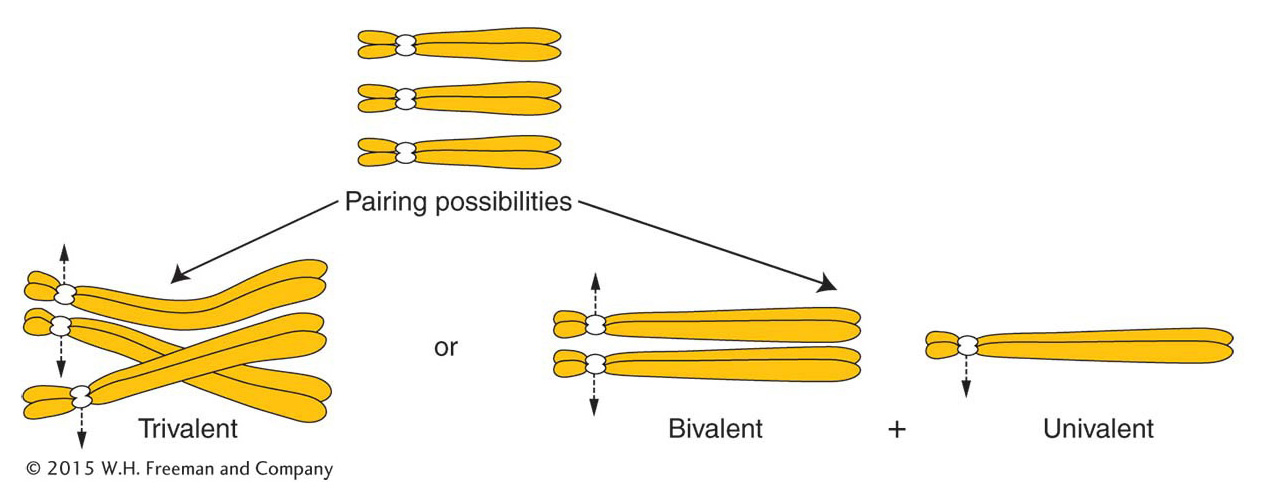
621
Aneuploid gametes do not generally give rise to viable offspring. In plants, aneuploid pollen grains are generally inviable and hence unable to fertilize the female gamete. In any organism, zygotes that might arise from the fusion of a haploid and an aneuploid gamete will themselves be aneuploid, and typically these zygotes also are inviable. We will examine the underlying reason for the inviability of aneuploids when we consider gene balance later in the chapter.
KEY CONCEPT
Polyploids with odd numbers of chromosome sets, such as triploids, are sterile or highly infertile because their gametes and offspring are aneuploid.Autotetraploids arise by the doubling of a 2n complement to 4n. This doubling can occur spontaneously, but it can also be induced artificially by applying chemical agents that disrupt microtubule polymerization. As stated in Chapter 2, chromosome segregation is powered by spindle fibers, which are polymers of the protein tubulin. Hence, disruption of microtubule polymerization blocks chromosome segregation. The chemical treatment is normally applied to somatic tissue during the formation of spindle fibers in cells undergoing division. The resulting polyploid tissue (such as a polyploid branch of a plant) can be detected by examining stained chromosomes from the tissue under a microscope. Such a branch can be removed and used as a cutting to generate a polyploid plant or allowed to produce flowers, which, when selfed, would produce polyploid offspring. A commonly used antitubulin agent is colchicine, an alkaloid extracted from the autumn crocus. In colchicine-
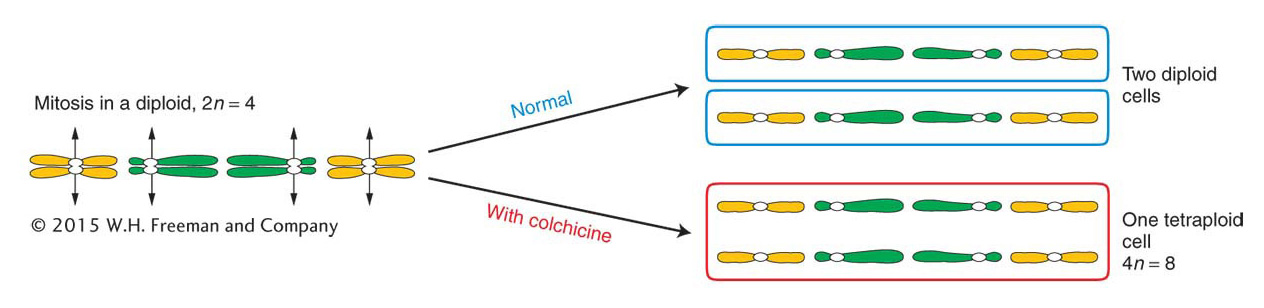
622
Because four is an even number, autotetraploids can have a regular meiosis, although this result is by no means always the case. The crucial factor is how the four chromosomes of each set pair and segregate. There are several possibilities, as shown in Figure 17-6. If the chromosomes pair as bivalents or quadrivalents, the chromosomes segregate normally, producing diploid gametes. The fusion of gametes at fertilization regenerates the tetraploid state. If trivalents form, segregation leads to nonfunctional aneuploid gametes and, hence, sterility.
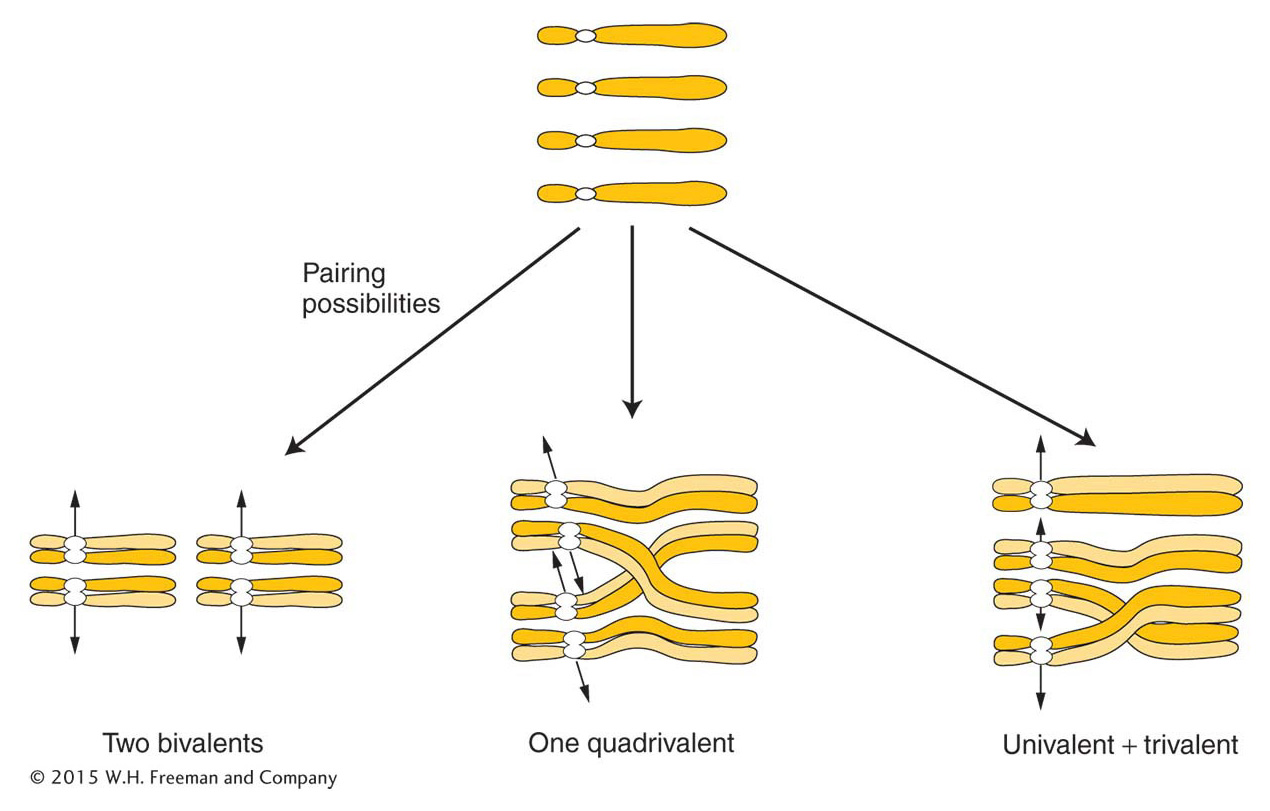
ANIMATED ART: Autotetraploid meiosis
623
What genetic ratios are produced by an autotetraploid? Assume for simplicity that the tetraploid forms only bivalents. If we start with an A/A/a/a tetraploid plant and self it, what proportion of progeny will be a/a/a/a? We first need to deduce the frequency of a/a gametes because this type is the only one that can produce a recessive homozygote. The a/a gametes can arise only if both pairings are A with a, and then both of the a alleles must segregate to the same pole. Let’s use the following thought experiment to calculate the frequencies of the possible outcomes. Consider the options from the point of view of one of the a chromosomes faced with the options of pairing with the other a chromosome or with one of the two A chromosomes; if pairing is random, there is a two-
KEY CONCEPT
If polyploids undergo orderly two-Allopolyploids An allopolyploid is a plant that is a hybrid of two or more species, containing two or more copies of each of the input genomes. The prototypic allopolyploid was an allotetraploid synthesized by Georgi Karpechenko in 1928. He wanted to make a fertile hybrid that would have the leaves of the cabbage (Brassica) and the roots of the radish (Raphanus), because they were the agriculturally important parts of each plant. Each of these two species has 18 chromosomes, and so 2n1 = 2n2 = 18, and n1 = n2 = 9. The species are related closely enough to allow intercrossing. Fusion of an n1 and an n2 gamete produced a viable hybrid progeny individual of constitution n1 + n2 = 18. However, this hybrid was functionally sterile because the 9 chromosomes from the cabbage parent were different enough from the radish chromosomes that pairs did not synapse and segregate normally at meiosis, and thus the hybrid could not produce functional gametes.
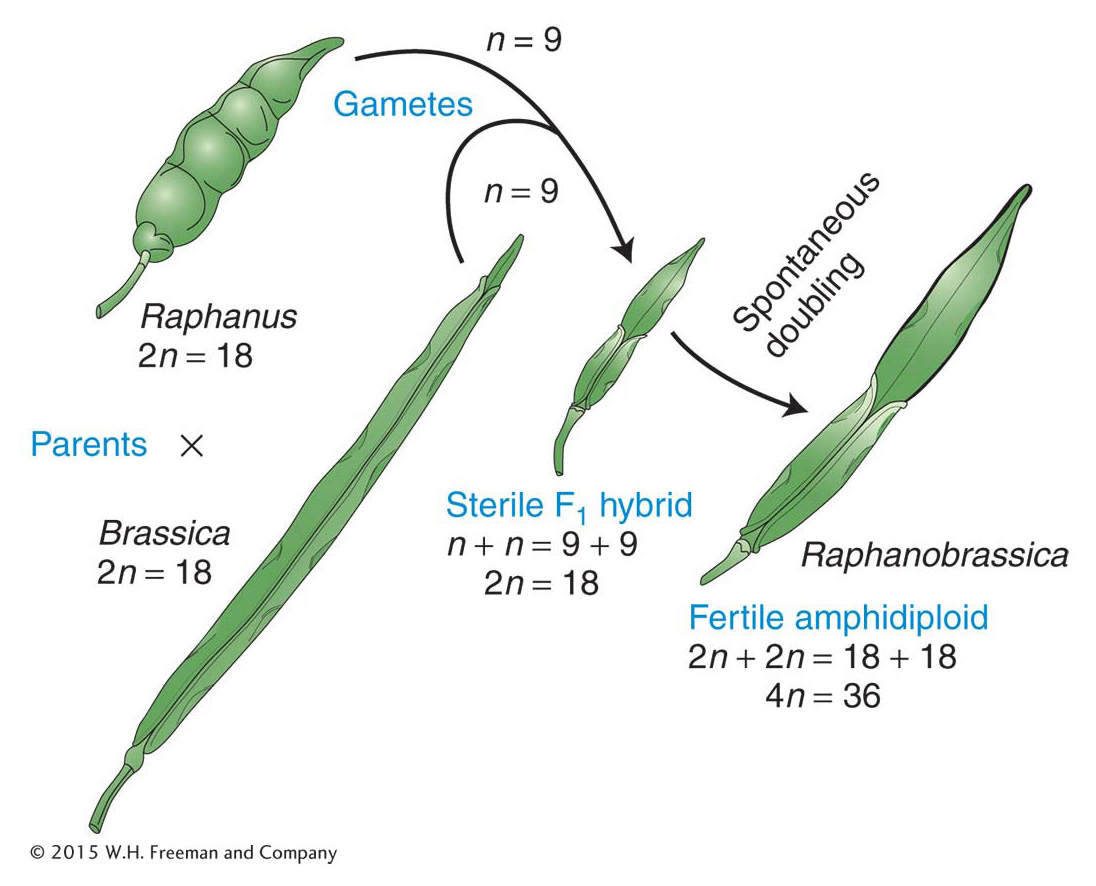
Eventually, one part of the hybrid plant produced some seeds. On planting, these seeds produced fertile individuals with 36 chromosomes. All these individuals were allopolyploids. They had apparently been derived from spontaneous, accidental chromosome doubling to 2n1 + 2n2 in one region of the sterile hybrid, presumably in tissue that eventually became a flower and underwent meiosis to produce gametes. In 2n1 + 2n2 tissue, there is a pairing partner for each chromosome, and functional gametes of the type n1 + n2 are produced. These gametes fuse to give 2n1 + 2n2 allopolyploid progeny, which also are fertile. This kind of allopolyploid is sometimes called an amphidiploid, or doubled diploid (Figure 17-7). Treating a sterile hybrid with colchicine greatly increases the chances that the chromosome sets will double. Amphidiploids are now synthesized routinely in this manner. (Unfortunately for Karpechenko, his amphidiploid had the roots of a cabbage and the leaves of a radish.)
When Karpechenko’s allopolyploid was crossed with either parental species—
624
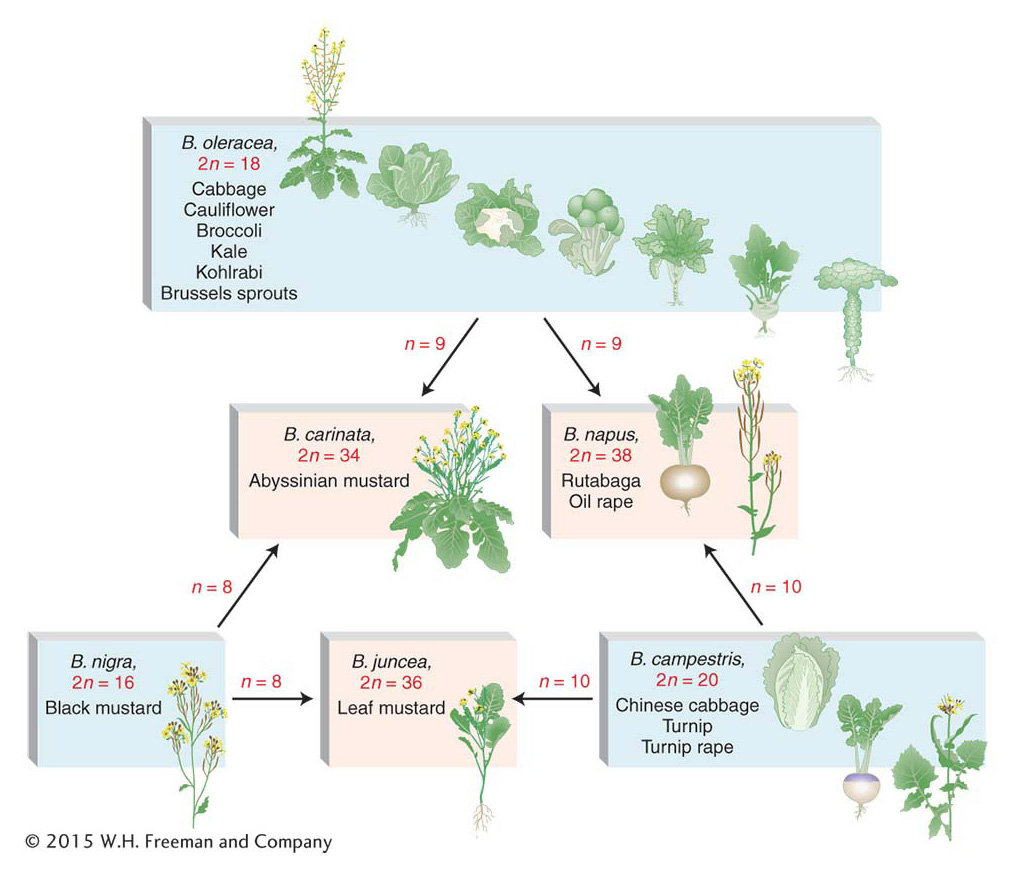
In nature, allopolyploidy seems to have been a major force in the evolution of new plant species. One convincing example is shown by the genus Brassica, as illustrated in Figure 17-8. Here, three different parent species have hybridized in all possible pair combinations to form new amphidiploid species. Natural polyploidy was once viewed as a somewhat rare occurrence, but recent work has shown that it is a recurrent event in many plant species. The use of DNA markers has made it possible to show that polyploids in any population or area that appear to be the same are the result of many independent past fusions between genetically distinct individuals of the same two parental species. An estimated 50 percent of all angiosperm plants are polyploids, resulting from auto-
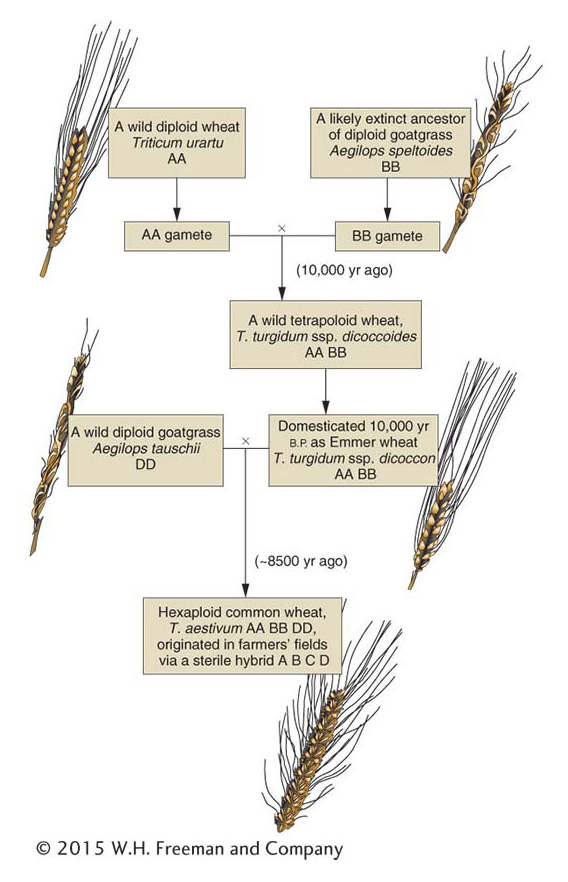
A particularly interesting natural allopolyploid is bread wheat, Triticum aestivum (6n = 42). By studying its wild relatives, geneticists have reconstructed a probable evolutionary history of this plant. Figure 17-9 shows that bread wheat is composed of two sets each of three ancestral genomes. At meiosis, pairing is always between homologs from the same ancestral genome. Hence, in bread-
625
Allopolyploid plant cells can also be produced artificially by fusing diploid cells from different species. First, the walls of two diploid cells are removed by treatment with an enzyme, and the membranes of the two cells fuse and become one. The nuclei often fuse, too, resulting in the polyploid. If the cell is nurtured with the appropriate hormones and nutrients, it divides to become a small allopolyploid plantlet, which can then be transferred to soil.
KEY CONCEPT
Allopolyploid plants can be synthesized by crossing related species and doubling the chromosomes of the hybrid or by fusing diploid cells.626
Agricultural applications Variations in chromosome number have been exploited to create new plant lines with desirable features. Some examples follow.
Monoploids Diploidy is an inherent nuisance for plant breeders. When they want to induce and select new recessive mutations that are favorable for agricultural purposes, the new mutations cannot be detected unless they are homozygous. Breeders may also want to find favorable new combinations of alleles at different loci, but such favorable allele combinations in heterozygotes will be broken up by recombination at meiosis. Monoploids provide a way around some of these problems.
Monoploids can be artificially derived from the products of meiosis in a plant’s anthers. A haploid cell destined to become a pollen grain can instead be induced by cold treatment (subjected to low temperatures) to grow into an embryoid, a small dividing mass of monoploid cells. The embryoid can be grown on agar to form a monoploid plantlet, which can then be potted in soil and allowed to mature (Figure 17-10).
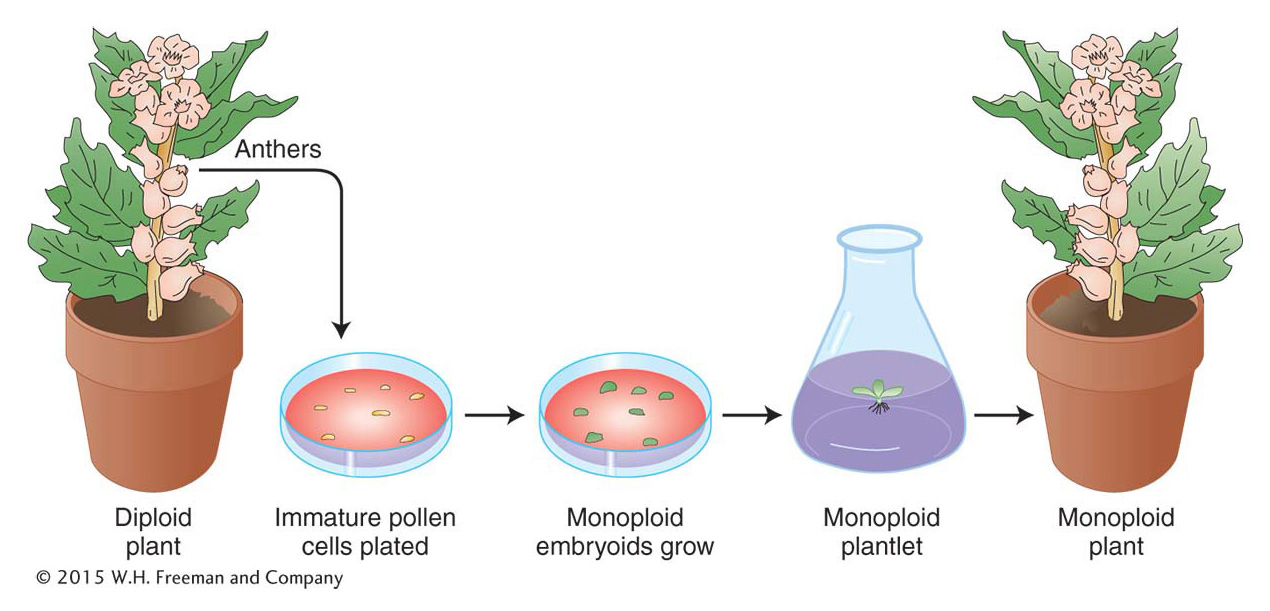
Plant monoploids can be exploited in several ways. In one approach, they are first examined for favorable allelic combinations that have arisen from the recombination of alleles already present in a heterozygous diploid parent. Hence, from a parent that is A/a; B/b might come a favorable monoploid combination a; b. The monoploid can then be subjected to chromosome doubling to produce homozygous diploid cells, a/a; b/b, that are capable of normal reproduction.
Another approach is to treat monoploid cells basically as a population of haploid organisms in a mutagenesis-
627
KEY CONCEPT
Geneticists can create new plant lines by producing I monoploids with favorable genotypes and then doubling their chromosomes to form fertile, homozygous diploids.Autotriploids The bananas that are widely available commercially are sterile triploids with 11 chromosomes in each set (3n = 33). The most obvious expression of the sterility of bananas is the absence of seeds in the fruit that we eat. (The black specks in bananas are not seeds; banana seeds are rock hard—
Autotetraploids Many autotetraploid plants have been developed as commercial crops to take advantage of their increased size (Figure 17-11). Large fruits and flowers are particularly favored.

Allopolyploids Allopolyploidy (formation of polyploids between different species) has been important in the production of modern crop plants. New World cotton is a natural allopolyploid that arose spontaneously, as is wheat. Allopolyploids are also synthesized artificially to combine the useful features of parental species into one type. Only one synthetic amphidiploid has ever been widely used commercially, a crop known as Triticale. It is an amphidiploid between wheat (Triticum, 6n = 42) and rye (Secale, 2n = 14). Hence, for Triticale, 2n = 2 × (21 + 7) = 56. This novel plant combines the high yields of wheat with the ruggedness of rye.
Polyploid animals As noted earlier, polyploidy is more common in plants than in animals, but there are cases of naturally occurring polyploid animals. Polyploid species of flatworms, leeches, and brine shrimps reproduce by parthenogenesis. Triploid and tetraploid Drosophila have been synthesized experimentally. However, examples are not limited to these so-
The sterility of triploids has been commercially exploited in animals as well as in plants. Triploid oysters have been developed because they have a commercial advantage over their diploid relatives. The diploids go through a spawning season, when they are unpalatable, but the sterile triploids do not spawn and are palatable year-
Aneuploidy
Aneuploidy is the second major category of chromosomal aberrations in which the chromosome number is abnormal. An aneuploid is an individual organism whose chromosome number differs from the wild type by part of a chromosome set. Generally, the aneuploid chromosome set differs from the wild type by only one chromosome or by a small number of chromosomes. An aneuploid can have a chromosome number either greater or smaller than that of the wild type. Aneuploid nomenclature (see Table 17-1) is based on the number of copies of the specific chromosome in the aneuploid state. For autosomes in diploid organisms, the aneuploid 2n + 1 is trisomic, 2n – 1 is monosomic, and 2n – 2 (the “– 2” represents the loss of both homologs of a chromosome) is nullisomic. In haploids, n + 1 is disomic. Special notation is used to describe sex-
628
Nondisjunction The cause of most aneuploidy is nondisjunction in the course of meiosis or mitosis. Disjunction is another word for the normal segregation of homologous chromosomes or chromatids to opposite poles at meiotic or mitotic divisions. Nondisjunction is a failure of this process, in which two chromosomes or chromatids incorrectly go to one pole and none to the other.
Mitotic nondisjunction can occur as cells divide during development. Sections of the body will be aneuploid (aneuploid sectors) as a result. Meiotic nondisjunction is more commonly encountered. In this case, the products of meiosis are aneuploid, leading to descendants in which the entire organism is aneuploid. In meiotic nondisjunction, the chromosomes may fail to disjoin at either the first or the second meiotic division (Figure 17-12). Either way, n – 1 and n + 1 gametes are produced. If an n – 1 gamete is fertilized by an n gamete, a monosomic (2n – 1) zygote is produced. The fusion of an n + 1 and an n gamete yields a trisomic 2n + 1.

ANIMATED ART: Meiotic nondisjunction
KEY CONCEPT
Aneuploid organisms result mainly from nondisjunction in a parental meiosis.Nondisjunction occurs spontaneously. Like most gene mutations, it is an example of a chance failure of a basic cellular process. The precise molecular processes that fail are not known but, in experimental systems, the frequency of nondisjunction can be increased by interference with microtubule polymerization, thereby inhibiting normal chromosome movement. Disjunction appears to be more likely to go awry in meiosis I. This failure is not surprising, because normal anaphase I disjunction requires that the homologous chromatids of the tetrad remain paired during prophase I and metaphase I, and it requires crossovers. In contrast, proper disjunction at anaphase II or at mitosis requires that the centromere split properly but does not require chromosome pairing or crossing over.
Crossovers are a necessary component of the normal disjunction process. Somehow the formation of a chiasma helps to hold a bivalent together and ensures that the two dyads will go to opposite poles. In most organisms, the amount of crossing over is sufficient to ensure that all bivalents will have at least one chiasma per meiosis. In Drosophila, many of the nondisjunctional chromosomes seen in disomic (n + 1) gametes are nonrecombinant, showing that they arise from meioses in which there is no crossing over on that chromosome. Similar observations have been made in human trisomies. In addition, in several different experimental organisms, mutations that interfere with recombination have the effect of massively increasing the frequency of meiosis I nondisjunction. All these observations provide evidence for the role of crossing over in maintaining chromosome pairing; in the absence of these associations, chromosomes are vulnerable to anaphase I nondisjunction.
629
KEY CONCEPT
Crossovers are needed to keep bivalents paired until anaphase I. If crossing over fails for some reason, first-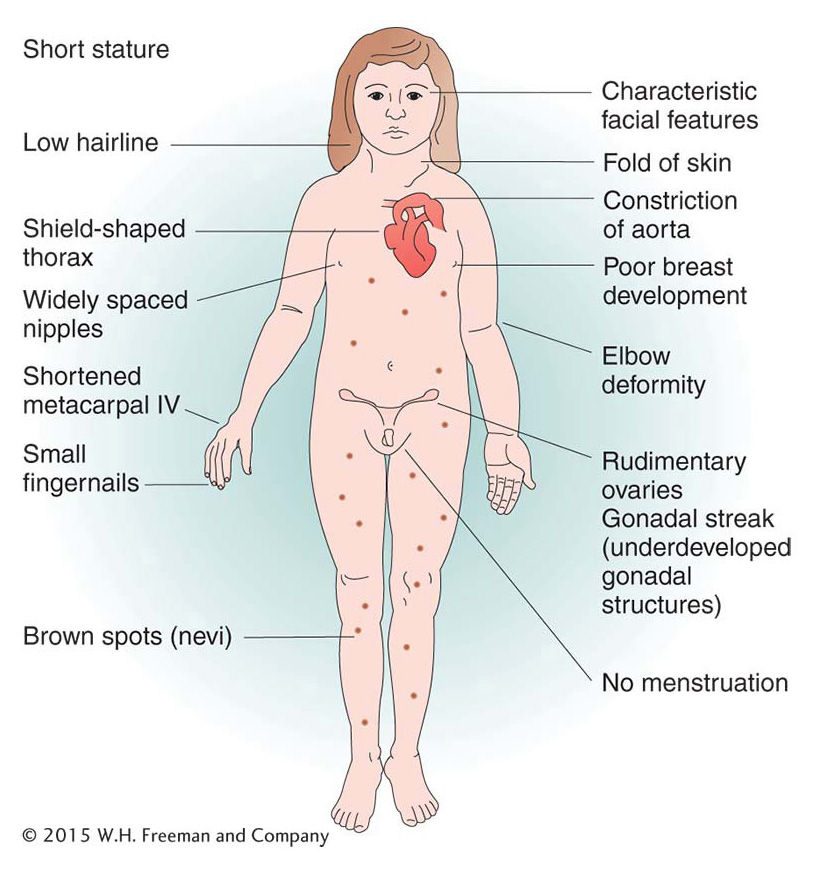
Monosomics (2n – 1) Monosomics are missing one copy of a chromosome. In most diploid organisms, the absence of one chromosome copy from a pair is deleterious. In humans, monosomics for any of the autosomes die in utero. Many X-
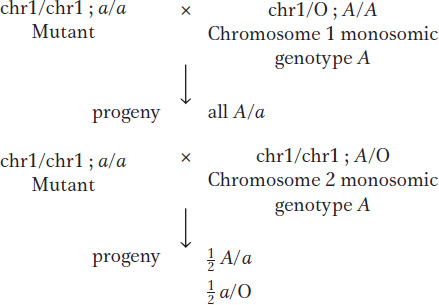
Geneticists have used viable plant monosomics to map newly discovered recessive mutant alleles to a specific chromosome. For example, one can make a set of monosomic lines, each known to lack a different chromosome. Homozygotes for the new mutant allele are crossed with each monosomic line, and the progeny of each cross are inspected for the recessive phenotype. The appearance of the recessive phenotype identifies the chromosome that has one copy missing as the one on which the gene is normally located. The test works because half the gametes of a fertile 2n – 1 monosomic will be n – 1, and, when an n – 1 gamete is fertilized by a gamete bearing a new mutation on the homologous chromosome, the mutant allele will be the only allele of that gene present and hence will be expressed.
As an illustration, let’s assume that a gene A/a is on chromosome 2. Crosses of a/a and monosomics for chromosome 1 and chromosome 2 are predicted to produce different results (chromosome 1 is abbreviated chr1):
Trisomics (2n + 1) Trisomics contain an extra copy of one chromosome. In diploid organisms generally, the chromosomal imbalance from the trisomic condition can result in abnormality or death. However, there are many examples of viable trisomics. Furthermore, trisomics can be fertile. When cells from some trisomic organisms are observed under the microscope at the time of meiotic chromosome pairing, the trisomic chromosomes are seen to form an associated group of three (a trivalent), whereas the other chromosomes form regular bivalents.
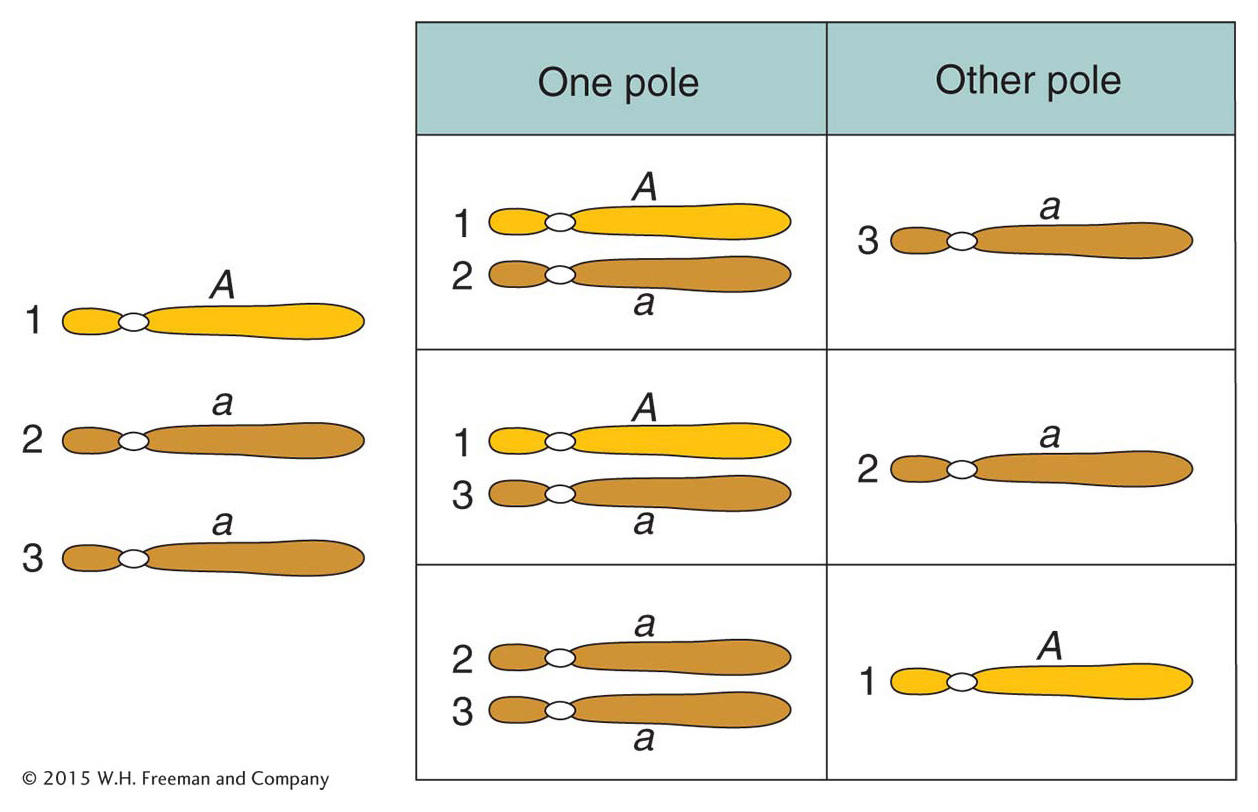
What genetic ratios might we expect for genes on the trisomic chromosome? Let’s consider a gene A that is close to the centromere on that chromosome, and let’s assume that the genotype is A/a/a. Furthermore, let’s postulate that, at anaphase I, the two paired centromeres in the trivalent pass to opposite poles and that the other centromere passes randomly to either pole. Then we can predict the three equally frequent segregations shown in Figure 17-14. These segregations result in an overall gametic ratio as shown in the six compartments of Figure 17-14; that is,
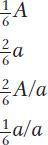
630
If a set of lines is available, each carrying a different trisomic chromosome, then a gene mutation can be located to a chromosome by determining which of the lines gives a trisomic ratio of the preceding type.
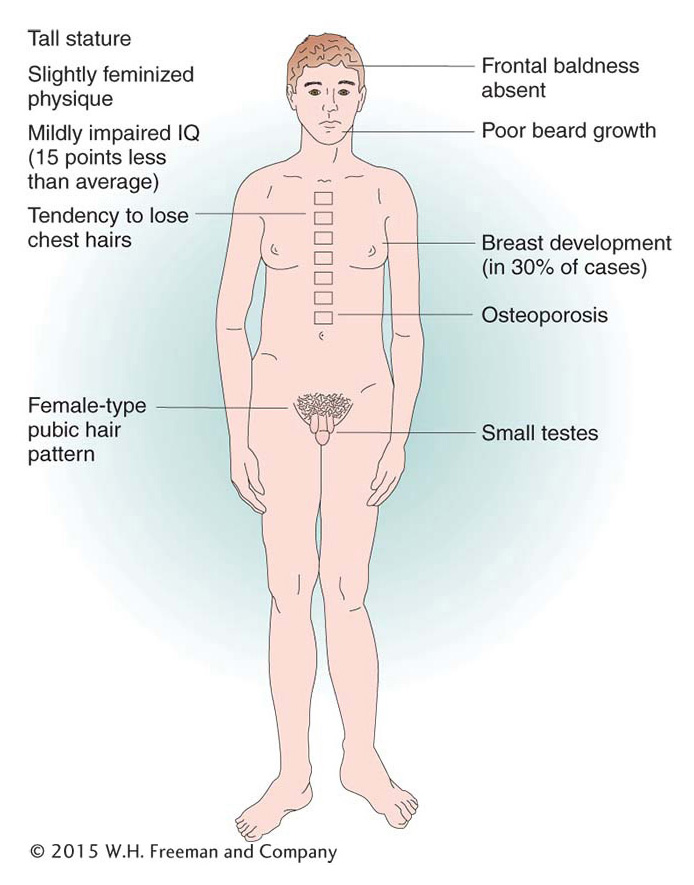
There are several examples of viable human trisomies. Several types of sex-
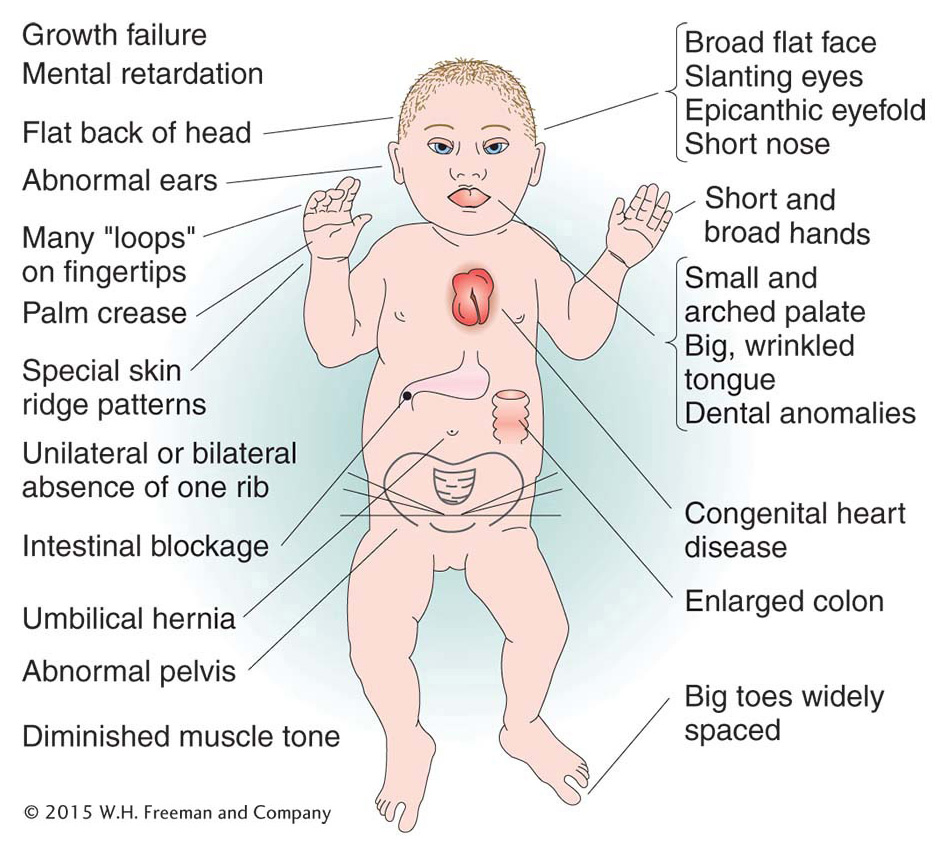
Of human trisomies, the most familiar type is Down syndrome (Figure 17-16), discussed briefly at the beginning of the chapter. The frequency of Down syndrome is about 0.15 percent of all live births. Most affected persons have an extra copy of chromosome 21 caused by nondisjunction of chromosome 21 in a parent who is chromosomally normal. In this sporadic type of Down syndrome, there is no family history of aneuploidy. Some rarer types of Down syndrome arise from translocations (a type of chromosomal rearrangement discussed later in the chapter); in these cases, as we will see, Down syndrome recurs in the pedigree because the translocation may be transmitted from parent to child.
The combined phenotypes that make up Down syndrome include mental retardation (with an IQ in the 20 to 50 range); a broad, flat face; eyes with an epicanthic fold; short stature; short hands with a crease across the middle; and a large, wrinkled tongue. Females may be fertile and may produce normal or trisomic progeny, but males are sterile with very few exceptions. Mean life expectancy is about 17 years, and only 8 percent of persons with Down syndrome survive past age 40.
631
The incidence of Down syndrome is related to maternal age: older mothers run a greatly elevated risk of having a child with Down syndrome (Figure 17-17). For this reason, fetal chromosome analysis (by amniocentesis or by chorionic villus sampling) is now recommended for older expectant mothers. A less-
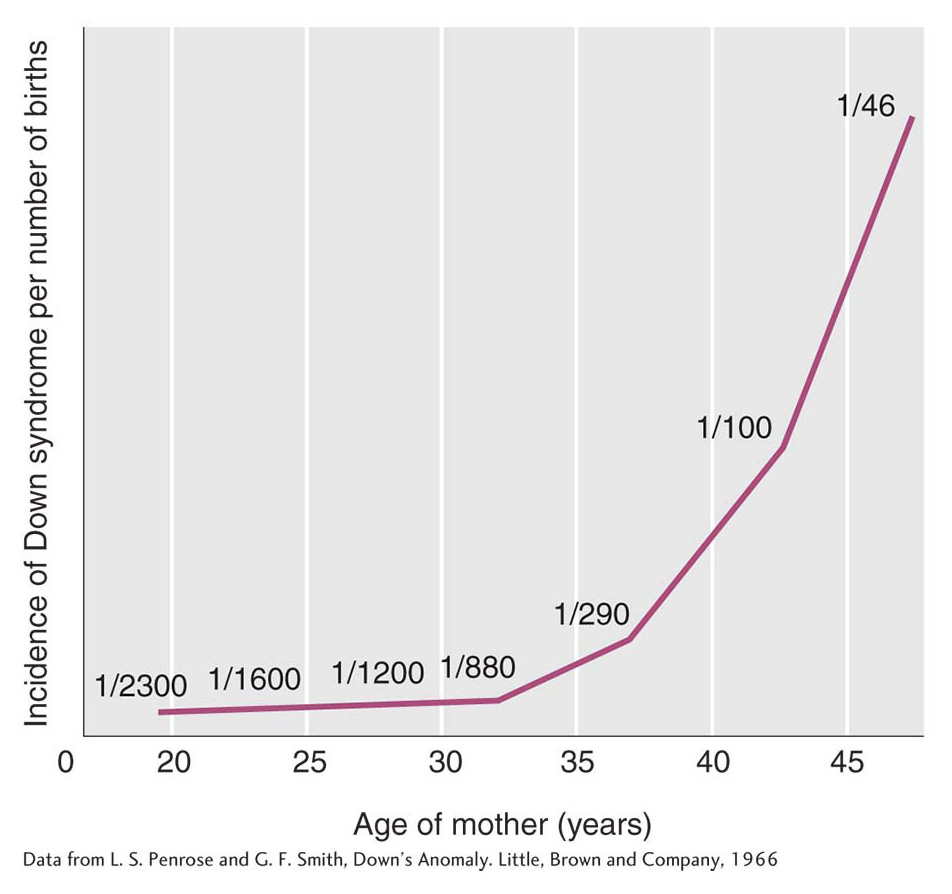
Even though the maternal-
The only other human autosomal trisomics to survive to birth are those with trisomy 13 (Patau syndrome) and trisomy 18 (Edwards syndrome). Both have severe physical and mental abnormalities. The phenotypic syndrome of trisomy 13 includes a harelip; a small, malformed head; “rocker-
632
The concept of gene balance
In considering aberrant euploidy, we noted that an increase in the number of full chromosome sets correlates with increased organism size but that the general shape and proportions of the organism remain very much the same. In contrast, autosomal aneuploidy typically alters the organism’s shape and proportions in characteristic ways.
Plants tend to be somewhat more tolerant of aneuploidy than are animals. Studies in jimsonweed (Datura stramonium) provide a classic example of the effects of aneuploidy and polyploidy. In jimsonweed, the haploid chromosome number is 12. As expected, the polyploid jimsonweed is proportioned like the normal diploid, only larger. In contrast, each of the 12 possible trisomics is disproportionate but in ways different from one another, as exemplified by changes in the shape of the seed capsule (Figure 17-18). The 12 different trisomies lead to 12 different and characteristic shape changes in the capsule. Indeed, these characteristics and others of the individual trisomics are so reliable that the phenotypic syndrome can be used to identify plants carrying a particular trisomy. Similarly, the 12 monosomics are themselves different from one another and from each of the trisomics. In general, a monosomic for a particular chromosome is more severely abnormal than is the corresponding trisomic.
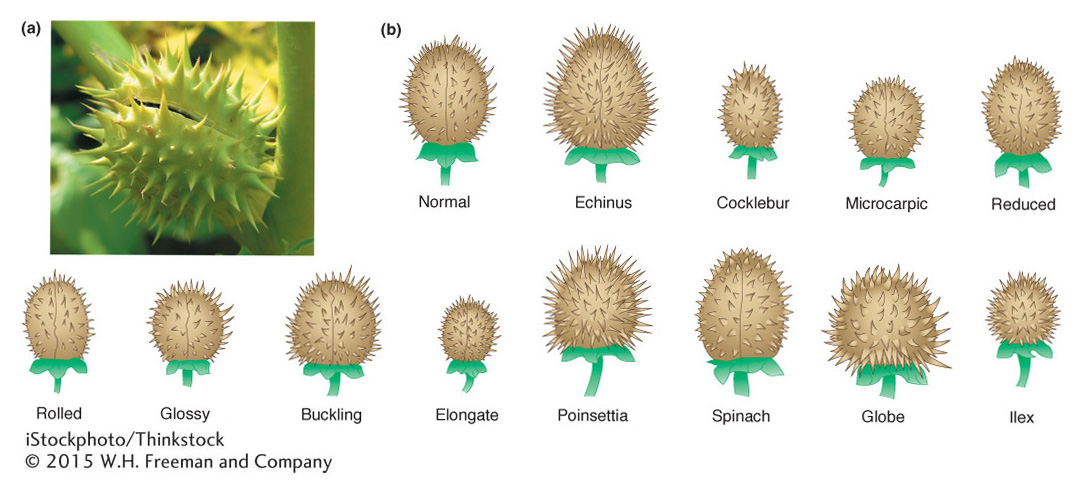
We see similar trends in aneuploid animals. In the fruit fly Drosophila, the only autosomal aneuploids that survive to adulthood are trisomics and monosomics for chromosome 4, which is the smallest Drosophila chromosome, representing only about 1 to 2 percent of the genome. Trisomics for chromosome 4 are only very mildly affected and are much less abnormal than are monosomics for chromosome 4. In humans, no autosomal monosomic survives to birth, but, as already stated, three types of autosomal trisomics can do so. As is true of aneuploid jimsonweed, each of these three trisomics shows unique phenotypic syndromes because of the special effects of altered dosages of each of these chromosomes.
Why are aneuploids so much more abnormal than polyploids? Why does aneuploidy for each chromosome have its own characteristic phenotypic effects? And why are monosomics typically more severely affected than are the corresponding trisomics? The answers seem certain to be a matter of gene balance. In a euploid, the ratio of genes on any one chromosome to the genes on other chromosomes is always 1:1, regardless of whether we are considering a monoploid, diploid, triploid, or tetraploid. For example, in a tetraploid, for gene A on chromosome 1 and gene B on chromosome 2, the ratio is 4 A:4 B, or 1:1. In contrast, in an aneuploid, the ratio of genes on the aneuploid chromosome to genes on the other chromosomes differs from the wild type by 50 percent: 50 percent for monosomics; 150 percent for trisomics. Using the same example as before, in a trisomic for chromosome 2, we find that the ratio of the A and B genes is 2 A:3 B. Thus, we can see that the aneuploid genes are out of balance. How does their being out of balance help us answer the questions raised?
633
In general, the amount of transcript produced by a gene is directly proportional to the number of copies of that gene in a cell. That is, for a given gene, the rate of transcription is directly related to the number of DNA templates available. Thus, the more copies of the gene, the more transcripts are produced and the more of the corresponding protein product is made. This relation between the number of copies of a gene and the amount of the gene’s product made is called a gene-
We can infer that normal physiology in a cell depends on the proper ratio of gene products in the euploid cell. This ratio is the normal gene balance. If the relative dosage of certain genes changes—
In some cases, the imbalances of aneuploidy result from the effects of a few “major” genes whose dosage has changed, rather than from changes in the dosage of all the genes on a chromosome. Such genes can be viewed as haplo-
However, the concept of gene balance does not tell us why having too few gene products (monosomy) is much worse for an organism than having too many gene products (trisomy). In a parallel manner, we can ask why there are many more haplo-
How do we apply the idea of gene balance to cases of sex-
634
In a sense, X chromosomes are naturally aneuploid. In species with an XY sexdetermination system, females have two X chromosomes, whereas males have only one. Nonetheless, the X chromosome’s housekeeping genes are expressed to approximately equal extents per cell in females and in males. In other words, there is dosage compensation. How is this compensation accomplished? The answer depends on the organism. In fruit flies, the male’s X chromosome appears to be hyperactivated, allowing it to be transcribed at twice the rate of either X chromosome in the female. As a result, the XY male Drosophila has an X gene dosage equivalent to that of an XX female. In mammals, in contrast, the rule is that no matter how many X chromosomes are present, there is only one transcriptionally active X chromosome in each somatic cell. This rule gives the XX female mammal an X gene dosage equivalent to that of an XY male. Dosage compensation in mammals is achieved by X-
Why are XXY individuals abnormal at all, given that triplo-
Gene dosage is also important in the phenotypes of polyploids. Human polyploid zygotes do arise through various kinds of mistakes in cell division. Most die in utero. Occasionally, triploid babies are born, but none survive. This fact seems to violate the principle that polyploids are more normal than aneuploids. The explanation for this contradiction seems to lie with X-