20.3 Molecular Evolution: The Neutral Theory
Darwin and Wallace conceived of evolution largely as “changes in organisms brought about by natural selection.” Indeed, this is what most people think of as the meaning of “evolution.” However, a century after Darwin’s theory, as molecular biologists began to confront evolution at the level of proteins and DNA molecules, they encountered and identified another dimension of the evolutionary process, neutral molecular evolution, which did not involve natural selection. An understanding of neutral molecular evolution is crucial to grasping how genes change over time.
The development of the neutral theory
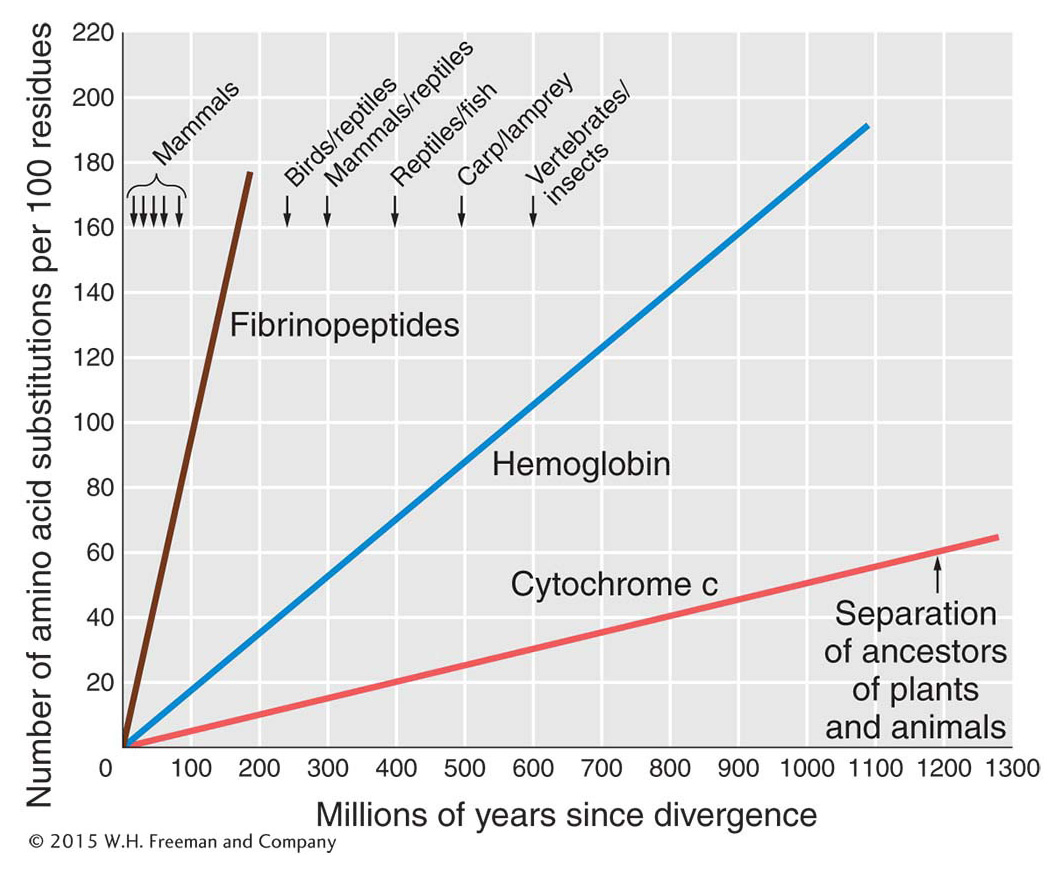
In the 1950s and early 1960s, methods were developed that enabled biologists to determine the amino acid sequences of proteins. This new capability raised the prospect that the fundamental basis of evolutionary change was finally at hand. However, as the sequences of proteins from a variety of species were deciphered, a paradox emerged. The sequences of globins and cytochrome c, for example, typically differ between any two species at a number of amino acids, and that number increases with the time elapsed since their divergence from a common ancestor (Figure 20-7). Yet, the function of these proteins is the same in different species—
The puzzle then was whether the amino acid replacements between species reflected changes in protein function and adaptations to selective conditions. Biochemists Linus Pauling and Emile Zuckerkandl did not think so. They observed that many substitutions were of one amino acid for another with similar properties. They concluded that most amino acid substitutions were “neutral” or “nearly neutral” and did not change the function of a protein whatsoever.
This line of reasoning was rejected at first by many evolutionary biologists, who at the time viewed all evolutionary changes as the result of natural selection and adaptation. Paleontologist George Gaylord Simpson argued that “there is a strong consensus that completely neutral genes or alleles must be rare if they exist at all. To an evolutionary biologist it therefore seems highly improbable that proteins … should change in a regular but non-
Zuckerkandl and Pauling asserted that the similarity or differences among organisms need not be reflected at the level of protein—
772
The debate was resolved by an onslaught of empirical data and the deciphering of the genetic code. Because multiple codons encode the same amino acid, a mutation that changes, say, CAG to CAC does not change the amino acid encoded. Therefore, variation can exist at the DNA level that has no effect on protein sequences, and thus neutral alleles do exist. But even more important for population genetics was the development of the “neutral theory of molecular evolution” by Motoo Kimura, Jack L. King, and Thomas Jukes. These authors proposed that most, but not all, mutations that are fixed are neutral or nearly neutral and any differences between species at such sites in DNA evolve by random genetic drift.
The “neutral theory” marked a profound conceptual shift away from a view of evolution as always guided by natural selection. Moreover, it provided a baseline assumption of how DNA should change over time if no other agent such as natural selection intervened.
6 G. G. Simpson, “Organisms and Molecules in Evolution,” Science 146, 1964, 1535–
KEY CONCEPT
The neutral theory of molecular evolution proposed that most mutations in DNA or amino acid replacements between species are functionally neutral or nearly neutral and fixed by random genetic drift. The assumption of neutrality offers a baseline expectation of how DNA should change over time when natural selection is absent.The rate of neutral substitutions
As we saw in Chapter 18 (see Box 18-5), we can calculate the expected rate of neutral changes in DNA sequences over time. If μ is the rate of new mutations at a locus per gene copy per generation, then the absolute number of new mutations that will appear in a population of N diploid individuals is 2Nμ. The new mutations are subject to random genetic drift: most will be lost from the population, while a few will become fixed and replace the previous allele. If a newly arisen mutation is neutral, then there is a probability of 1/(2N) that it will replace the previous allele because of random genetic drift. Each one of the 2Nμ new mutations that will appear in a population has a probability of 1/(2N) of eventually taking over that population. Thus, the absolute substitution rate k is the mutation rate multiplied by the probability that any one mutation will eventually take over by drift:
K = rate of neutral substitution = 2Nμ × 1/(2N) = μ
That is, we expect that, in every generation, there will be μ substitutions in the population, purely from the genetic drift of neutral mutations.
KEY CONCEPT
The rate of substitutions in DNA in evolution resulting from the random genetic drift of neutral mutations is equal to the mutation rate to such alleles, μ.The signature of purifying selection on DNA
When measurements of molecular change deviate from what is expected for neutral changes, that is an important signal—
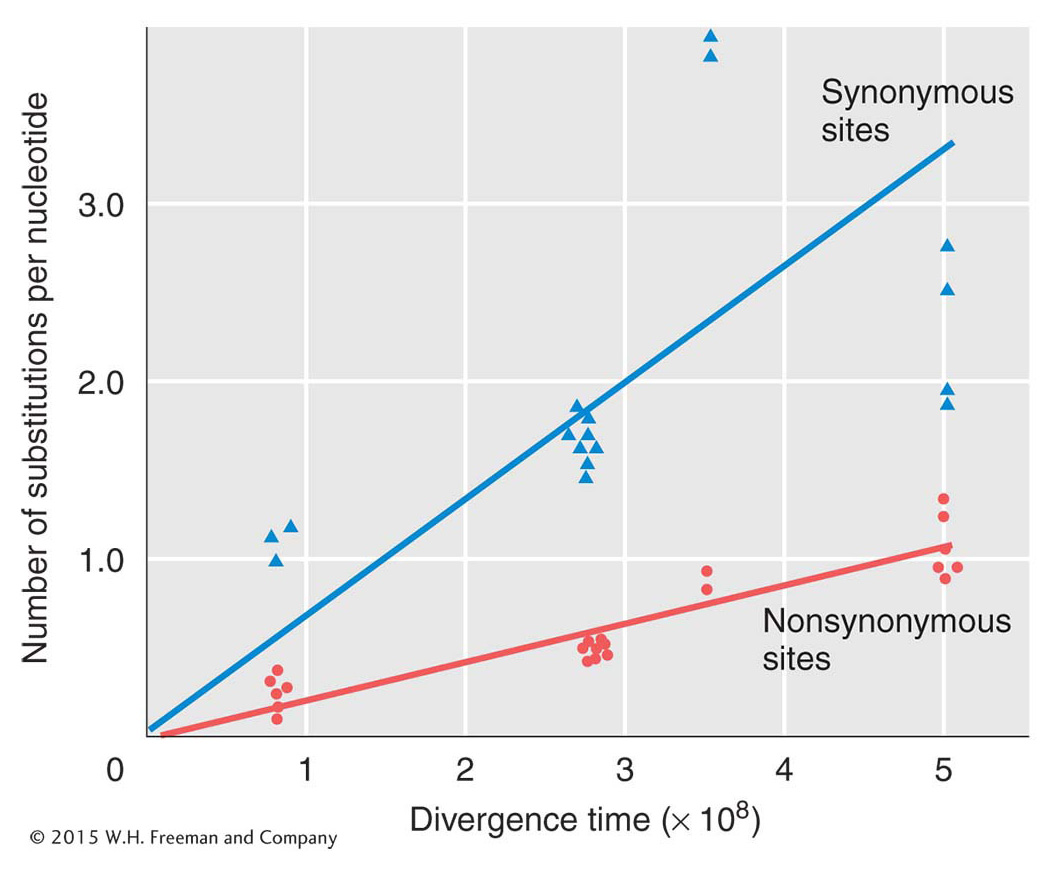
All classes of DNA sequences, including exons, introns, regulatory sequences, and sequences in between genes, show nucleotide diversity among individuals within populations and between species. The constant rate of neutral substitutions predicts that, if the number of nucleotide differences between two species were plotted against the time since their divergence from a common ancestor, the result should be a straight line with slope equal to μ. That is, evolution should proceed according to a molecular clock that is ticking at the rate μ. Figure 20-8 shows such a plot for the ß-globin gene. The results are quite consistent with the claim that nucleotide substitutions have been neutral in the past 500 million years. Two sorts of neutral nucleotide substitutions are plotted: synonymous substitutions, which are from one alternative codon to another, making no change in the amino acid, and nonsynonymous substitutions, which result in an amino acid change. Figure 20-8 shows a much lower slope for nonsynonymous substitutions than for synonymous changes, which means that the substitution rate of neutral nonsynonymous substitutions is much lower than that of synonymous neutral substitutions.
773
This outcome is precisely what we expect under natural selection. Mutations that cause an amino acid substitution should have a deleterious effect more often than synonymous substitutions, which do not change the protein. Such deleterious variants will be removed from populations by purifying selection (see Chapter 18). A lower-
Purifying selection is the most widespread, but often overlooked, facet of natural selection. The “rejection of injurious variations,” as Darwin termed it, is pervasive. Purifying selection explains why we find many protein sequences that are unchanged or nearly unchanged over vast spans of evolutionary time. For example, there are several dozen genes that exist in all domains of life—
KEY CONCEPT
Purifying selection is a pervasive aspect of natural selection that reduces genetic variation and preserves DNA and protein sequences over aeons of time.Another prediction of the theory of neutral evolution is that different proteins will have different clock rates, because the metabolic functions of some proteins will be much more sensitive to changes in their amino acid sequences. Proteins in which every amino acid makes a difference will have a lower rate of neutral mutation because a smaller proportion of their mutations will be neutral compared with proteins that are more tolerant of substitution. Figure 20-7 shows a comparison of the clocks for fibrinopeptides, hemoglobin, and cytochrome c. That fibrinopeptides have a much higher proportion of neutral mutations is reasonable because these peptides are merely a nonmetabolic safety catch, cut out of fibrino-
KEY CONCEPT
The rate of neutral evolution for the amino acid sequence of a protein depends on the sensitivity of the protein’s function to amino acid changes.774
The conservation of gene sequences by purifying selection and the neutral evolution of gene sequences are two crucial dimensions of the evolutionary process, but neither of them account for the origin of adaptations. In the next two sections of the chapter, we will illustrate several examples of the ways in which genetic changes are linked to changes in traits and to organismal diversity.