Molecular structure determines enzyme function
Many enzymes are much larger than their substrates. An enzyme is typically a protein containing hundreds of amino acids. It may consist of a single folded polypeptide chain or of several subunits (see Key Concept 3.2). Its substrate is generally a small molecule or a small part of a large molecule. The active site of the enzyme is usually quite small, not more than 6 to 12 amino acids. You may be asking yourself:
What features of the enzyme’s active site allow it to recognize and bind the substrate?
What is the role of the rest of the protein?
THE ACTIVE SITE IS SPECIFIC TO THE SUBSTRATE(S) The remarkable ability of an enzyme to select exactly the right substrate(s) depends on a precise interlocking of molecular shapes and interactions of chemical groups at the active site. The binding of a substrate to the active site depends on the same kinds of forces that maintain the tertiary structure of the enzyme: hydrogen bonds, the attraction and repulsion of electrically charged groups, and hydrophobic interactions. The specific fit of the substrate in the active site of lysozyme is illustrated in Figures 8.9 and 8.11B.
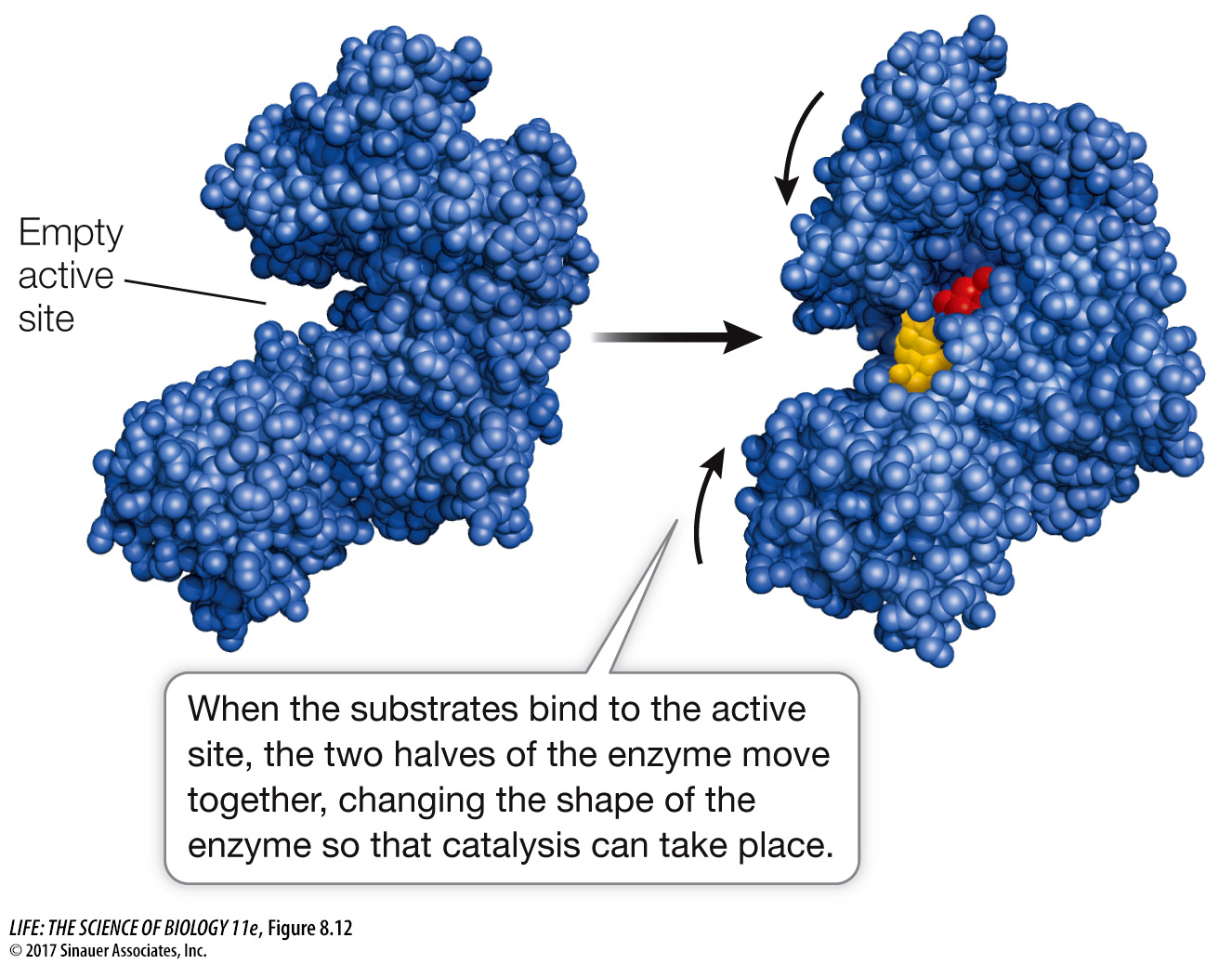
AN ENZYME CHANGES SHAPE WHEN IT BINDS A SUBSTRATE Just as a membrane receptor protein may undergo precise changes in conformation upon binding to its ligand (see Chapter 7), some enzymes change their shapes when they bind their substrate(s). These shape changes, which are called induced fit, alter the shape of the active site(s) of the enzyme.
An example of induced fit can be seen in the enzyme hexokinase (see Figure 8.7), which catalyzes the reaction
Glucose + ATP → glucose 6-
Induced fit brings reactive side chains from the hexokinase active site into alignment with the substrates (Figure 8.12), facilitating its catalytic mechanisms. Equally important, the folding of hexokinase to fit around the substrates (glucose and ATP) excludes water from the active site. This is essential, because if water were present, the ATP could be hydrolyzed to ADP and Pi. But since water is absent, the transfer of a phosphate from ATP to glucose is favored.
Question
Q: Do covalent bonds in hexokinase break when it changes shape? Explain.
No. Typically, the tertiary structure of a protein is due to weak noncovalent forces such as hydrophobic interactions, ionic attractions, and hydrogen bonds. Binding of substrate alters these forces but not covalent bonds.
162
Induced fit at least partly indicates what the role(s) of the rest of the protein may be:
It provides a framework so that the amino acids of the active site are properly positioned in relation to the substrate(s).
It participates in significant changes in protein shape and structure that result in induced fit.
It provides binding sites for regulatory molecules (see Key Concept 8.5).