Genes can be exchanged between chromatids and mapped
If linkage were absolute, Mendel’s law of independent assortment would apply only to loci on different chromosomes. Instead, genes at different loci on the same chromosome do sometimes separate from one another during meiosis. Genes may recombine when two homologous chromosomes physically exchange corresponding segments during prophase I of meiosis—
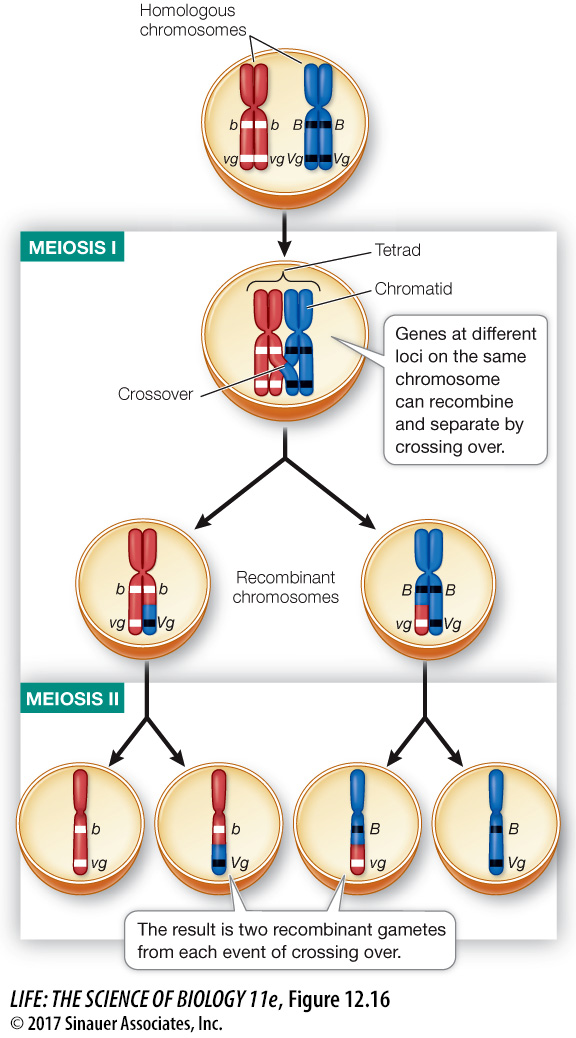
Note that one exchange event involves only two of the four chromatids in a tetrad, one from each member of the homologous pair, and can occur at any point along the length of the chromosome. The chromosome segments involved are exchanged reciprocally, so both chromatids involved in crossing over become recombinant (that is, each chromatid ends up with genes from both of the organism’s parents). Usually several exchange events occur along the length of each homologous pair.
When crossing over takes place between two linked genes, not all the progeny of a cross have the parental phenotypes. Instead, recombinant offspring appear as well, as they did in Morgan’s test cross (see Figure 12.15). They appear in proportions called recombinant frequencies, which are calculated by dividing the number of recombinant progeny by the total number of progeny (Figure 12.17). Recombinant frequencies will be greater for loci that are farther apart on the chromosome than for loci that are closer together because an exchange event is more likely to occur between genes that are far apart.
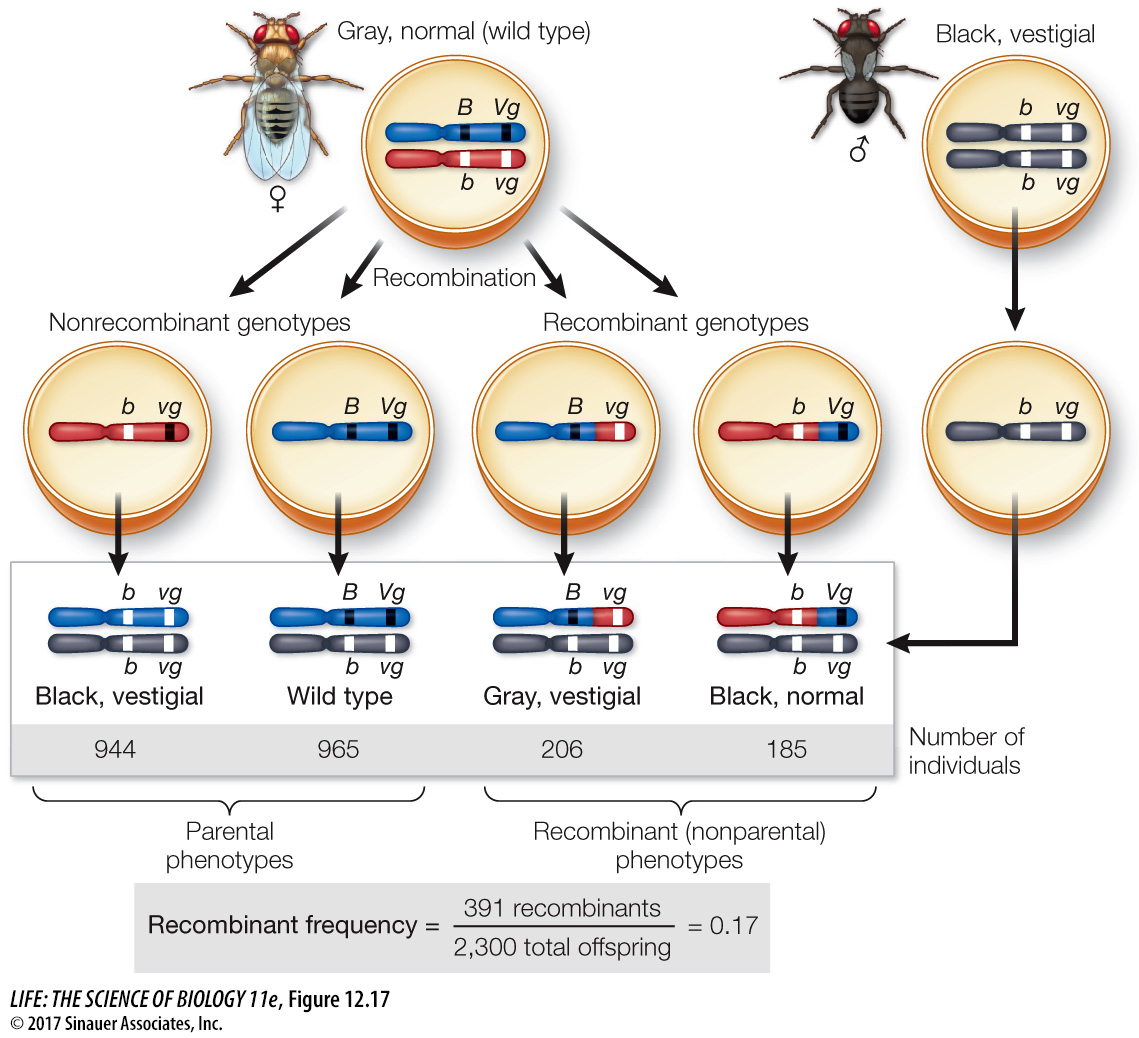
By calculating recombinant frequencies, geneticists can infer the locations of genes along a chromosome and generate a genetic map. Below is a map showing five genes on a fruit fly chromosome constructed using the recombination frequencies generated by test crosses involving various pairs of genes:
258
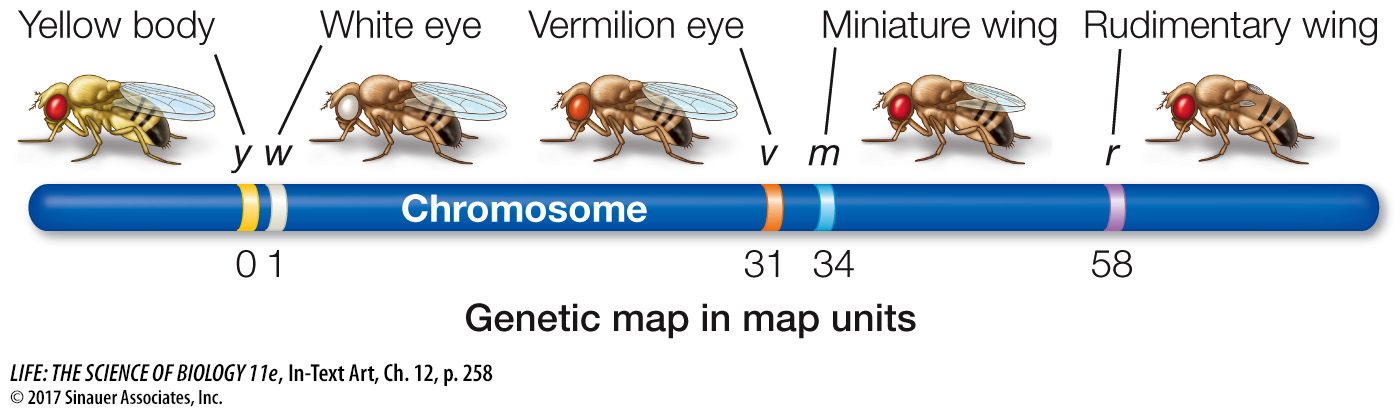
In the chromosome shown above, the recombination frequency between y and w is low, so they are close together on the map. Recombination between y and v is more frequent, so they are farther apart. The recombination frequencies are converted to map units (also called centiMorgans, cM); one map unit is equivalent to an average recombination frequency of 0.01 (1%).
*Gene sequencing of DNA has made chromosome mapping less important in some areas of genetics research. However, mapping is still a way to verify that a particular DNA sequence corresponds with a particular phenotype. Linkage has allowed biologists to isolate genes and to identify genetic markers linked to important genes. This is important in breeding new crops and animals for agriculture, and for identifying people carrying medically significant mutations.
*connect the concepts As described in Key Concept 17.1, gene sequencing techniques reveal the nucleotide sequences of genes and identify where genes begin and end on the chromosome.