DNA polymerases add nucleotides to the growing chain
DNA replication begins with the binding of a large protein complex (the pre-
276
investigating life
The Meselson–
experiment
Original Paper: Meselson, M. and F. Stahl. 1958. The replication of DNA in Escherichia coli. Proceedings of the National Academy of Sciences USA 44: 671–
A centrifuge was used to separate DNA molecules labeled with isotopes of different densities. This experiment revealed a pattern that supports the semiconservative model of DNA replication.
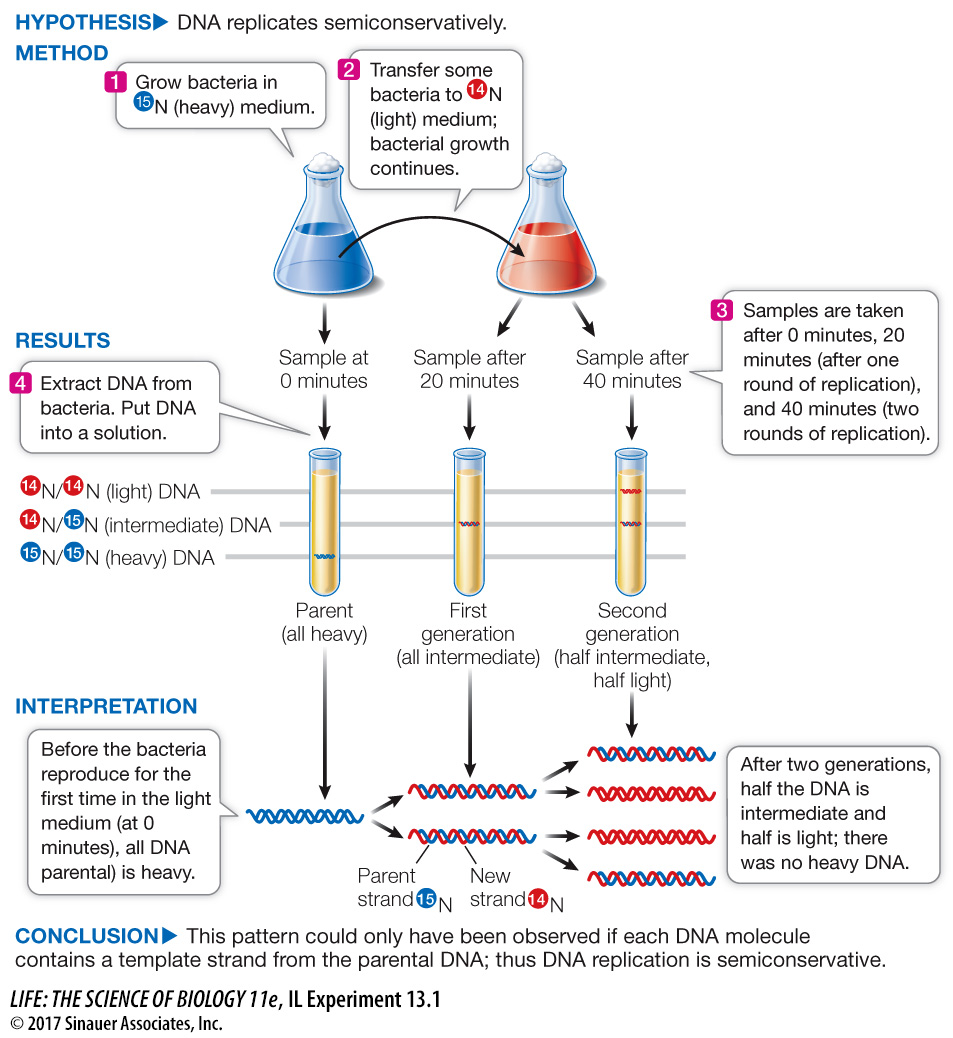
277
work with the data
The Meselson–
Figure B shows results of the experiment after each of four generations. Each sample contained the same number of bacteria, so the total amount of DNA in each panel was the same.
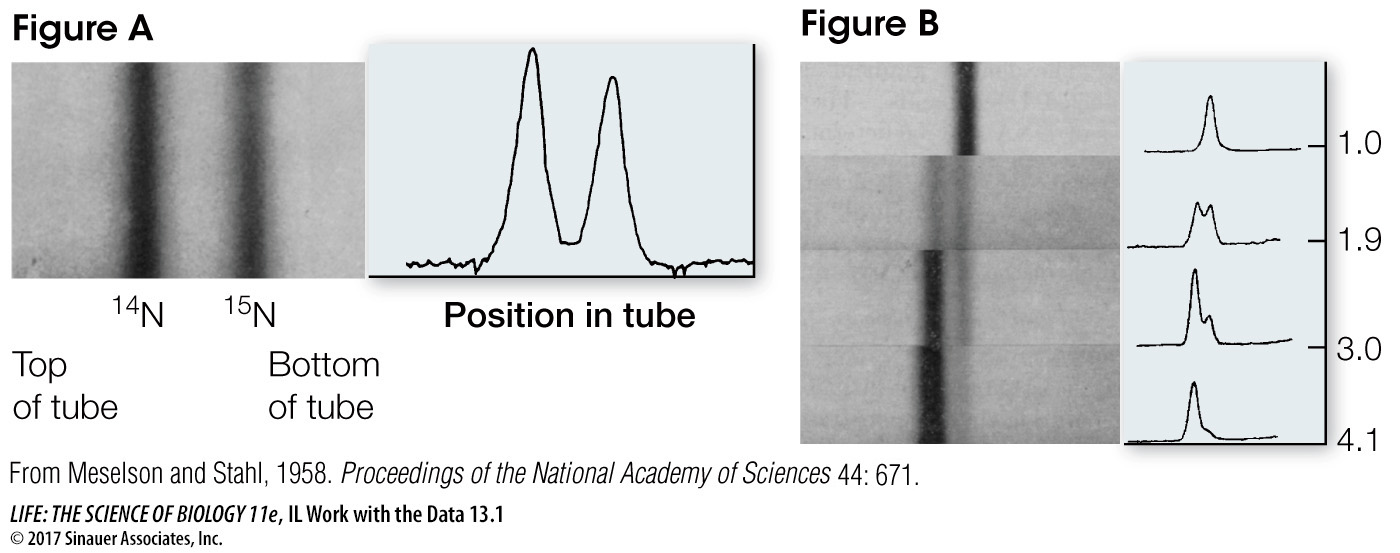
QUESTIONS
Question 1
Use the heights of the peaks to estimate the percent of total DNA that was heavy, intermediate, and light at each generational stage. Create a table summarizing these calculations and discuss whether they support the authors’ conclusions.
These data fit the semiconservative model of DNA replication because the heavy strands were templates for new light strands; after one round of replication, all the DNA had one heavy (original) strand and one light (newly made) strand, and so was intermediate in weight.
| Generation | Percent heavy DNA | Percent intermediate DNA | Percent light DNA |
|---|---|---|---|
| 1 | 0 | 100 | 0 |
| 2 | 0 | 50 | 50 |
| 3 | 0 | 25 | 75 |
| 4 | 0 | 12.5 | 87.5 |
Question 2
What would the data look like if the bacteria had been allowed to divide for three more generations?
After seven generations there would be about 1.5 percent intermediate DNA and 98.5 percent light DNA.
Question 3
If Meselson and Stahl had done their experiment starting with light DNA and then added 15N for succeeding generations, what would the bands look like?
In the first generation, the bands would be the same as in Figure B: all intermediate. In the second generation, 1/2 would be intermediate and 1/2 heavy. In the third generation, 1/4 would be intermediate and 3/4 heavy. In the fourth generation, 1/8 would be intermediate and 7/8 heavy.
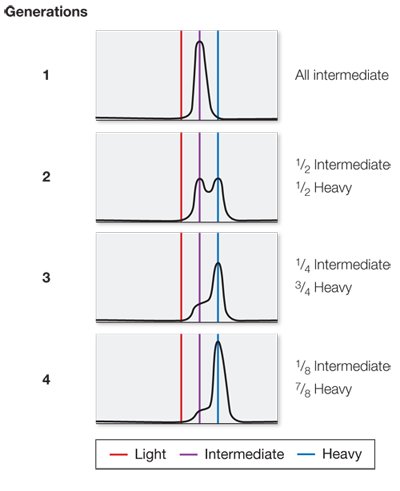
Question 4
What would the data look like if conservative replication were the correct model? What would the data look like if dispersive replication were correct?
In a conservative model, the first generation would be 1/2 heavy, 1/2 light; the second generation 1/4 heavy, 3/4 light; the third generation 1/8 heavy, 7/8 light; and the fourth generation 1/16 heavy, 15/16 light. In a dispersive model, the first generation would be all intermediate; the second generation all half-
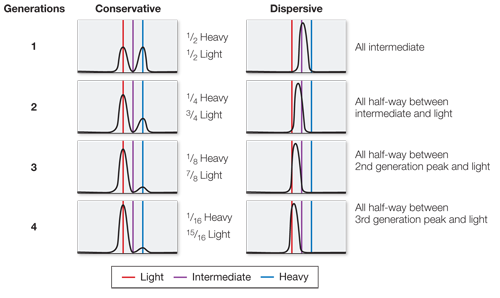
A similar work with the data exercise may be assigned in LaunchPad.
ORIGINS OF REPLICATION The single circular chromosome of the bacterium E. coli has 4 × 106 base pairs (bp) of DNA. The 245 bp ori sequence is at a particular location on the chromosome. Once the pre-
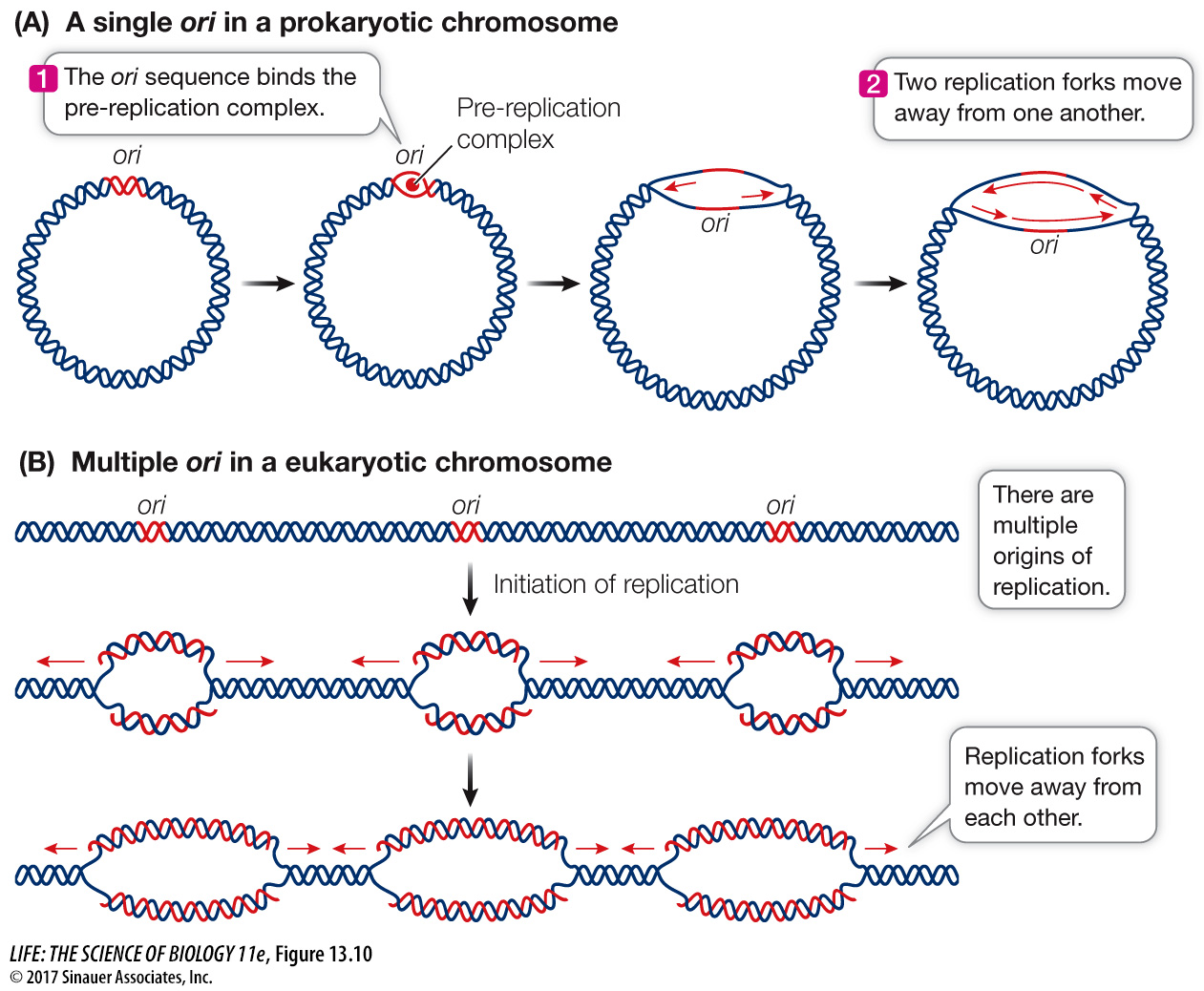
Eukaryotic chromosomes are typically much longer than those of prokaryotes—
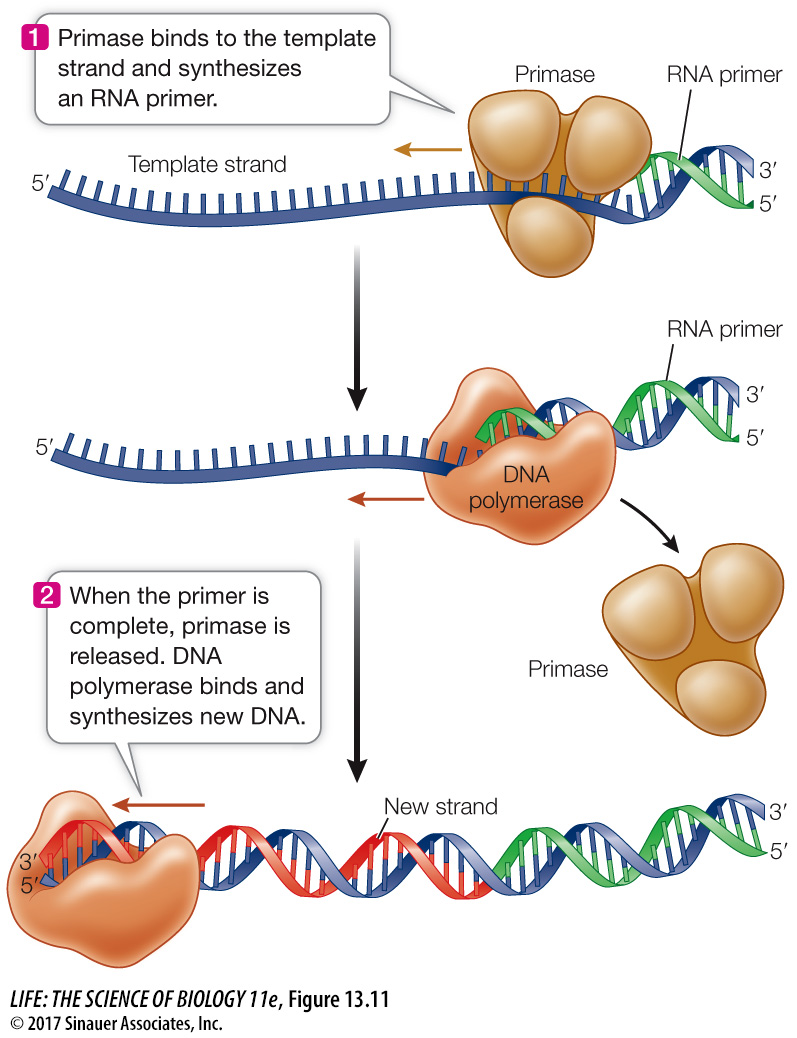
DNA REPLICATION BEGINS WITH A PRIMER A DNA polymerase elongates a polynucleotide strand by covalently linking new nucleotides to a preexisting strand. However, it cannot start this process without a short “starter” strand, called a primer. In most organisms this primer is a short single strand of RNA (Figure 13.11), but in some organisms it is DNA. The primer is complementary to the DNA template and is synthesized one nucleotide at a time by an enzyme called a primase. The DNA polymerase then adds nucleotides to the 3′ end of the primer and continues until the replication of that section of DNA has been completed. Then the RNA primer is degraded, DNA is added in its place, and the resulting DNA fragments are connected by the action of other enzymes. When DNA replication is complete, each new strand consists only of DNA.
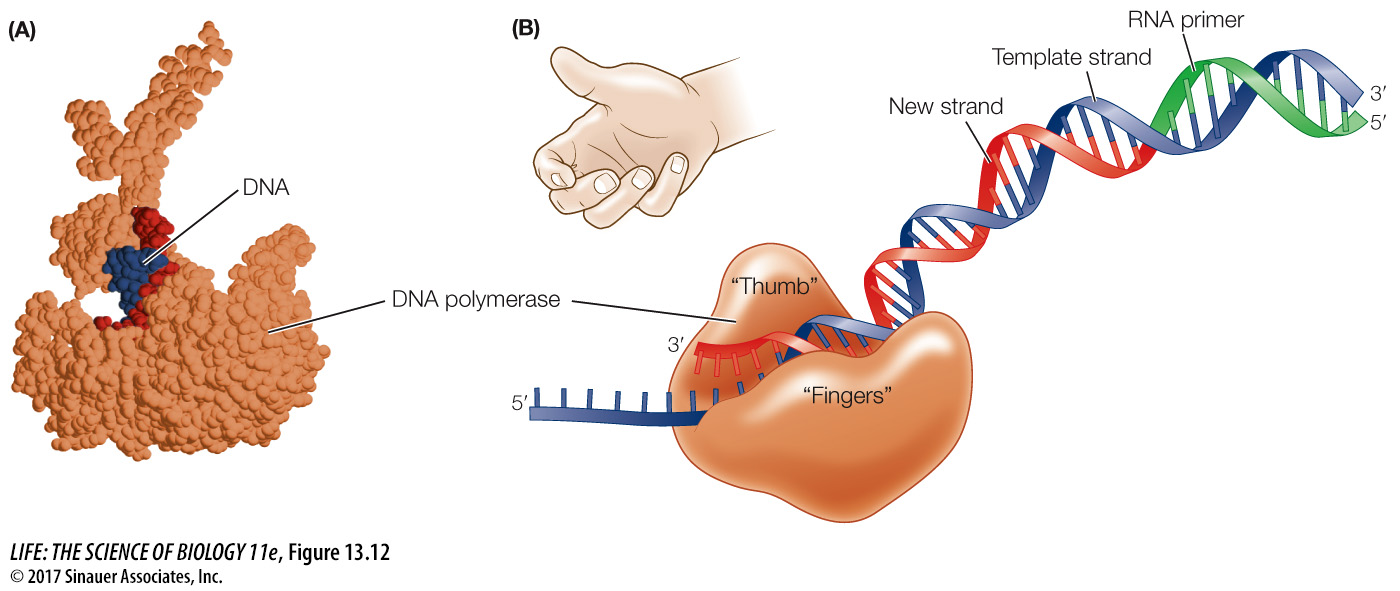
DNA POLYMERASES ARE LARGE DNA polymerases are much larger than their substrates (the dNTPs) and the template DNA, which is very thin. Molecular models of the enzyme–
278