The polymerase chain reaction can make multiple copies of a DNA sequence
In order to study DNA and perform genetic manipulations in the lab, it is often necessary to make multiple copies of a DNA sequence. DNA amplification is needed because the amount of DNA isolated from a biological sample is often too small to work with. The polymerase chain reaction (PCR) technique essentially automates this replication process by copying a short region of DNA many times in a test tube.
The PCR reaction mixture contains:
A sample of double-
stranded DNA from a biological sample, to act as the template Two short, artificially synthesized primers that are complementary to the ends of the sequence to be amplified
The four dNTPs (dATP, dTTP, dCTP, and dGTP)
A DNA polymerase that can tolerate high temperatures without becoming degraded
Salts and a buffer to maintain a near-
neutral pH
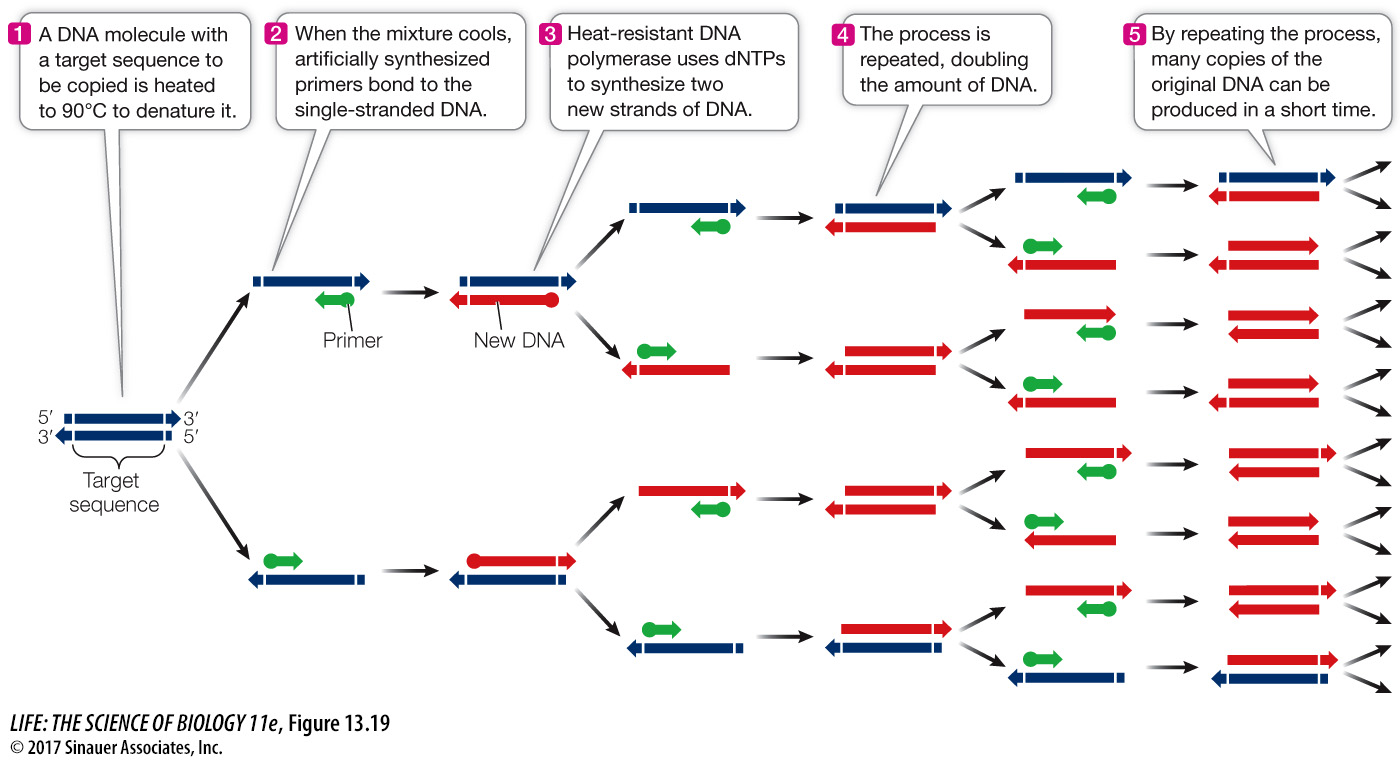
PCR amplification is a cyclic process in which a sequence of steps is repeated over and over (Figure 13.19):
Question
Q: How do PCR primers relate to DNA replication primers?
Both PCR primers in the test tube and DNA replication primers in the cell act to begin DNA replication. However, PCR primers can be complementary to any DNA strand, whereas DNA replication primers bind only to the origin of replication.
285
The first step involves heating the reaction mixture to near boiling point, to separate (denature) the two strands of the DNA template.
The reaction is then cooled to allow the primers to bind (or anneal) to the template strands.
Next, the reaction is warmed to an optimum temperature for the DNA polymerase to catalyze the production of the complementary new strands.
A single cycle takes a few minutes to produce two copies of the target DNA sequence, leaving the new DNA in the double-
The PCR technique requires that the base sequences at the 3′ end of each strand of the target DNA sequence be known, so that complementary primers, usually 15 to 30 bases long, can be made in the laboratory. Because of the uniqueness of DNA sequences, a pair of primers this length will usually bind to only a single region of DNA in an organism’s genome. This specificity, despite the incredible diversity of DNA sequences, is a key to the power of PCR.
One initial problem with PCR was its temperature requirements. To denature DNA, it must be heated to more than 90°C—
Scientists pondering the problem of copying DNA by PCR read Brock’s basic research articles and got a clever idea: why not use T. aquaticus DNA polymerase in the PCR technique? It could withstand the 90°C denaturation temperature and would not have to be added during each cycle. The idea worked, and it earned biochemist Kary Mullis a Nobel prize. PCR has had an enormous impact on genetic research. Some of its most striking applications will be described in Chapters 15–18. These applications range from amplifying DNA in order to identify an individual person or organism, to detection of diseases.