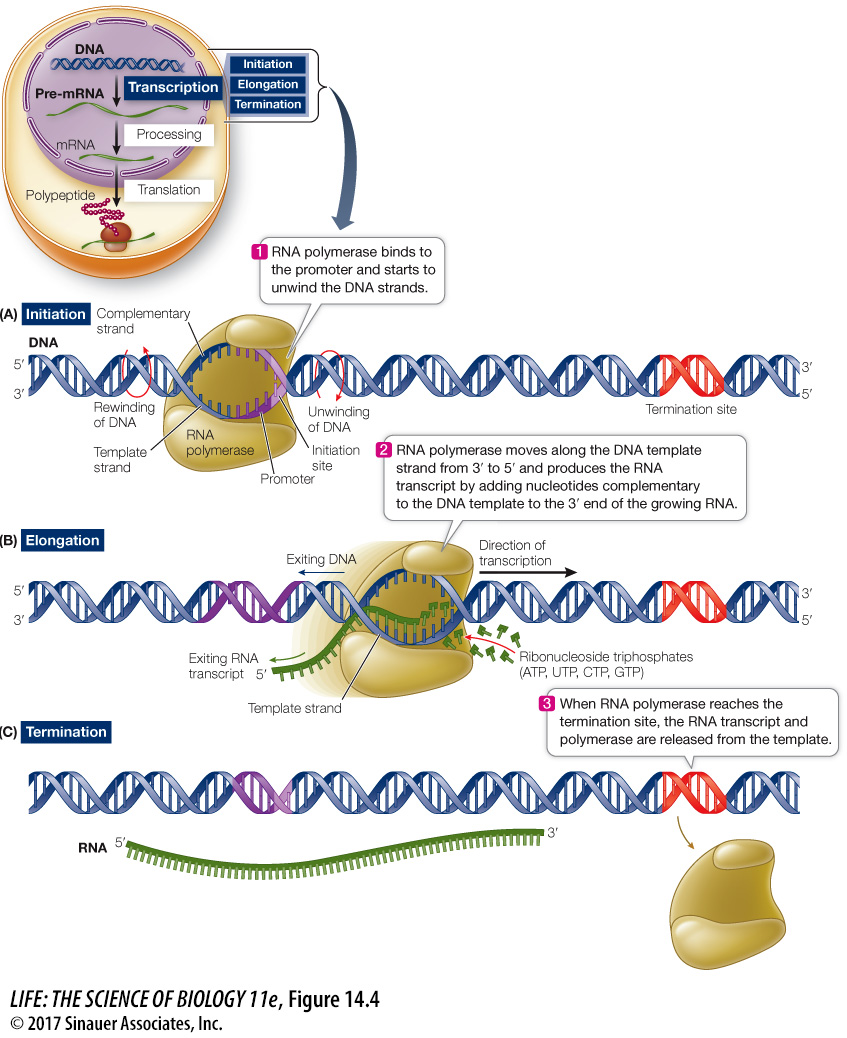
Transcription occurs in three steps
294
Transcription can be divided into three distinct processes: (1) initiation, (2) elongation, and (3) termination. Follow these processes in Figure 14.4.
Question
Q: Comparing RNA synthesis with DNA replication, what are the common features?
Both RNA synthesis and DNA replication require a polymerase enzyme that binds to DNA; the DNA must unwind to expose the bases; the new polymer is complementary to a template strand; and the substrates are nucleoside triphosphates.
Animation 14.1 Transcription
www.life11e.com/
INITIATION Transcription begins when RNA polymerase binds to a special sequence of DNA called a promoter (see Figure 14.4A). Eukaryotic genes generally have one promoter each, whereas in prokaryotes and viruses, several genes often share one promoter. Promoters are important control sequences that “tell” the RNA polymerase two things:
Where to start transcription
Which strand of DNA to transcribe
A promoter reads in a particular direction, so it orients the RNA polymerase and thus “aims” it at the appropriate strand to use as a template. Part of each promoter is the initiation site, where transcription begins. Groups of nucleotides lying “upstream” from the initiation site (5′ on the non-
Although every gene has a promoter, not all promoters are identical. Some are more effective at transcription initiation than others. Furthermore, there are differences between transcription initiation in prokaryotes and in eukaryotes. We will discuss promoters and their roles in the regulation of gene expression in Chapter 16.
ELONGATION After RNA polymerase has bound to the promoter, it begins the process of elongation (see Figure 14.4B). DNA unwinds about 10 base pairs at a time and RNA polymerase reads the template strand in the 3′-to-
You may recall from Key Concept 13.3 that DNA polymerase uses dNTPs (deoxyribonucleoside triphosphates) as substrates, and forms covalent bonds between each incoming dNTP and the 3′ end of the growing polynucleotide chain (see Figure 13.10). Energy released by the removal of two phosphate groups from the dNTP is used to drive the reaction. Similarly, RNA polymerase uses (ribo)nucleoside triphosphates (NTPs) as substrates, removing two phosphate groups from each substrate molecule and using the released energy to drive the polymerization reaction.
Because RNA polymerases do not proofread, transcription errors occur at a rate of one for every 104 to 105 bases. Because many copies of RNA are made, however, and because they often have only a relatively short life span, these errors are not as potentially harmful as mutations in DNA.
TERMINATION Just as initiation sites in the DNA template strand specify the starting point for transcription, particular base sequences specify its termination (see Figure 14.4C). There are two mechanisms for ending transcription. For some genes, the newly formed transcript folds back on itself and forms internal hydrogen bonds between bases. A loop forms, and this structure causes the transcript to fall away from the DNA template and the RNA polymerase. In other cases, a protein binds to specific sequences on the transcript and causes the RNA to detach from the DNA template.