Noncoding sequences called introns often appear between genes in eukaryotic chromosomes
Differences between prokaryotic and eukaryotic transcription are revealed when mRNA probes from prokaryotes and eukaryotes are incubated with their respective chromosomal DNAs:
In prokaryotes (Figure 14.6B, top), there is usually a 1:1 linear complementarity between the base sequence of the mRNA at the ribosome and that of the chromosomal DNA.
In eukaryotes (Figure 14.6B, bottom), one or more non-
hybridized DNA loops are often observed to extrude out of the mRNA– DNA hybrid, indicating that there are stretches of DNA sequence that do not have a complementary sequence in the mRNA that is translated at the ribosome.
300
The discovery that some stretches of eukaryotic DNA sequence are excluded from the mRNA that reaches the ribosome for translation initiated the question of whether this “extra” DNA actually gets transcribed. Does transcription “skip” these “extra” sequences, or are they transcribed and then somehow edited out of the mRNA transcript before it arrives at the ribosome? To find out, an experiment can be conducted in which the initial mRNA transcript in the cell nucleus—
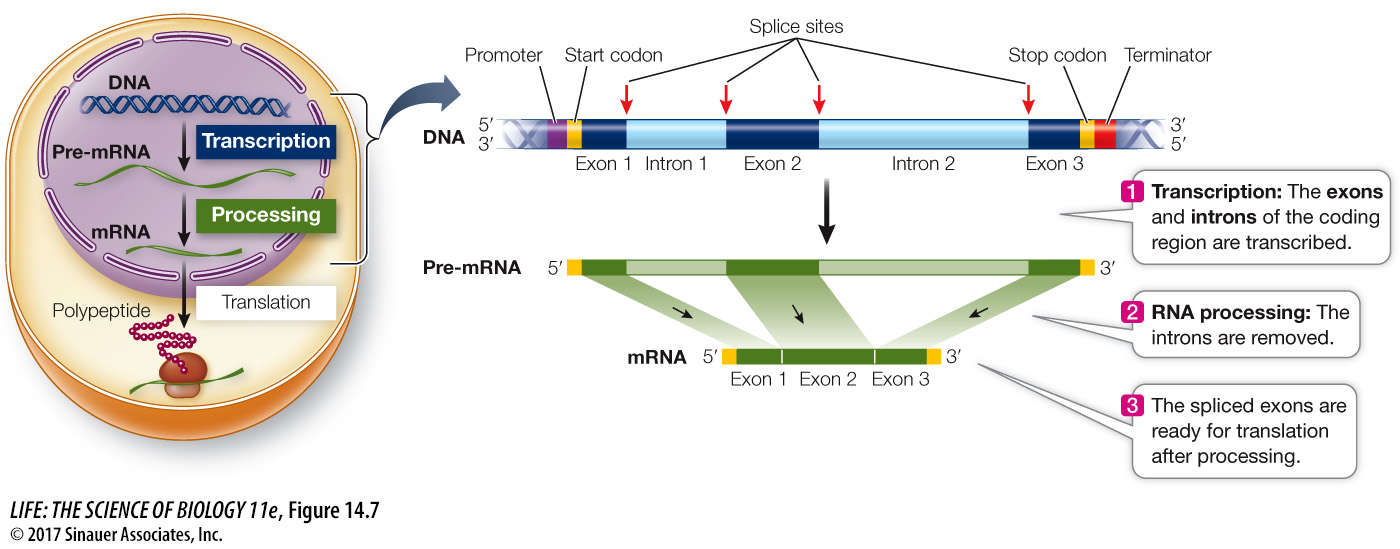
Introns interrupt, but do not scramble, the DNA sequence of a gene. The base sequences of the exons in the template strand, if joined and taken in order, form a continuous sequence that is complementary to that of the mature mRNA. In some cases, the separated exons often encode different functional regions, or domains, of the protein. For example, the globin polypeptides that make up hemoglobin each have two domains: one for binding to a nonprotein pigment called heme, and another for binding to the other globin subunits. These two domains are encoded by different exons in the globin genes. Most (but not all) eukaryotic genes contain introns, and in rare cases, introns are also found in prokaryotes. The largest human gene encodes a muscle protein called titin; it has 363 exons, which together code for 38,138 amino acids.