Pre-
The transcript of a eukaryotic gene is modified in several ways before it leaves the nucleus: both ends of the pre-
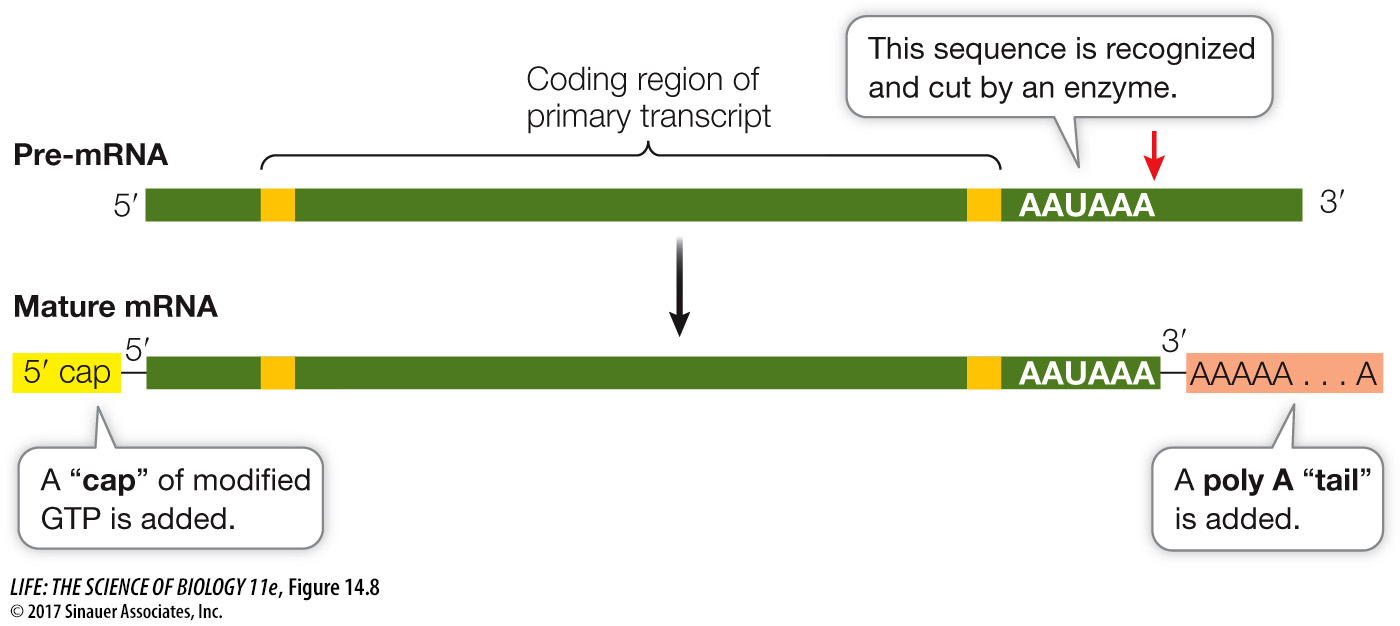
MODIFICATION AT BOTH ENDS Two steps in the processing of pre-
A 5′ cap is added to the 5′ end of the pre-
mRNA as it is transcribed. The 5′ cap, which is a chemically modified molecule of guanosine triphosphate (GTP), facilitates the binding of mRNA to the ribosome for translation, and it protects the mRNA from being digested by ribonucleases that break down RNAs. A poly A tail is added to the 3′ end of the pre-
mRNA at the end of transcription. Transcription ends downstream of the termination codon in DNA. In eukaryotes there is usually a “polyadenylation” sequence (AAUAAA) near the 3′ end of the pre- mRNA, after the last codon. This sequence acts as a signal for an enzyme to cut the pre- mRNA. Immediately after this cleavage, another enzyme adds 100 to 300 adenine nucleotides (the poly A tail) to the 3′ end of the pre- mRNA. This tail helps in the export of mature mRNA from the nucleus and is important for mRNA stability.
301
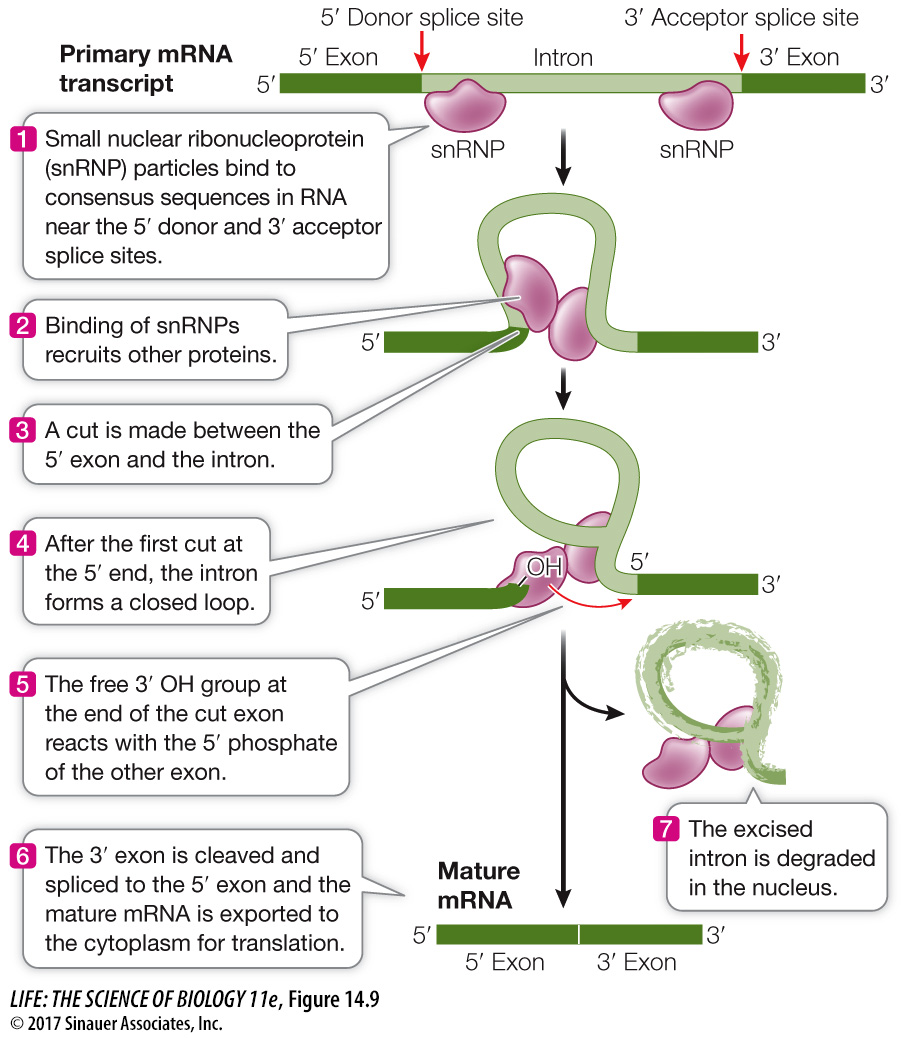
SPLICING TO REMOVE INTRONS Within the nucleus, introns are removed from eukaryotic pre-
Animation 14.3 RNA Splicing
www.life11e.com/
Molecular studies of human genetic diseases have provided insights into RNA splicing. For example, people with the genetic disease beta thalassemia have a defect in the production of one of the hemoglobin subunits. These people suffer from severe anemia because they have an inadequate supply of red blood cells. In some cases, the genetic mutation that causes the disease occurs at an intron consensus sequence, where the splicing machinery binds to the RNA (see Figure 14.9, step 1) in the β-globin gene. Consequently, β-globin pre-
After processing is completed in the nucleus, the mature mRNA moves out into the cytoplasm through the nuclear pores. A protein bound to the 5′ nucleotide cap during processing is recognized by a receptor at the nuclear pore. Together, these proteins lead the mRNA through the pore. Unprocessed or incompletely processed pre-