How are proteins directed to their cellular destinations?
As a polypeptide chain emerges from the ribosome it may simply fold into its three-
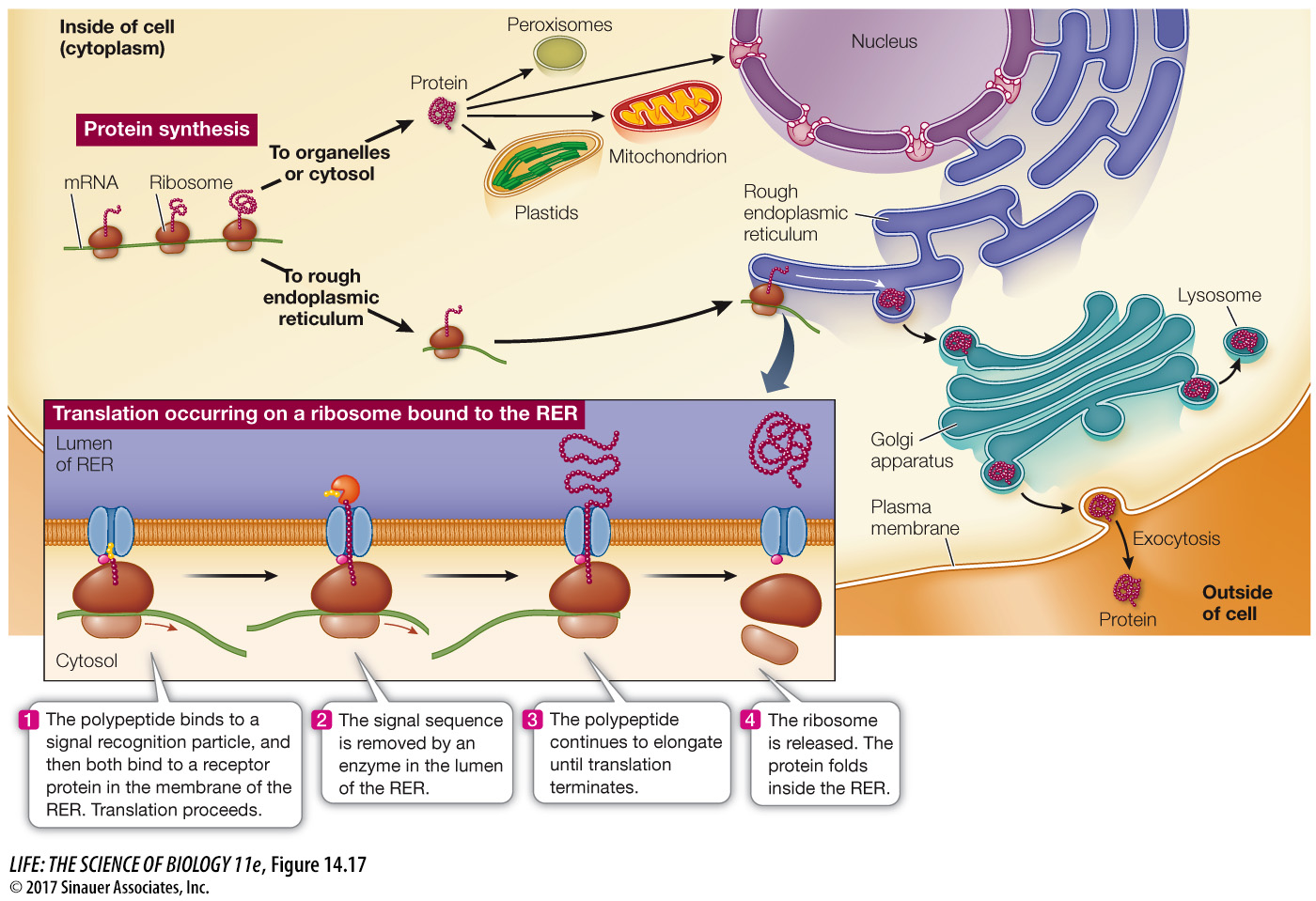
Protein synthesis always begins on free ribosomes, and the “default” location for a protein is the cytosol. In the absence of a signal sequence, the protein will remain in the same cellular compartment in which it was synthesized. Some proteins contain signal sequences that “target” them to the nucleus, mitochondria, plastids, or peroxisomes (Figure 14.17). A signal sequence binds to a specific receptor protein at the surface of the organelle. Once it has bound, the targeted protein moves into the organelle. For example, here is a nuclear localization signal (NLS):
-Pro-
Question
Q: What happens to a protein that has no amino acid sequence “address”?
A protein with no “address” stays in the cytoplasm.
How do we know this signal sequence directs the protein to the nucleus? The function of this NLS peptide was established using experiments like the one illustrated in Figure 14.18. Proteins were made in the laboratory with or without the peptide, and then tested by injecting them into cells. Only proteins with the NLS were found in the nucleus.
experiment
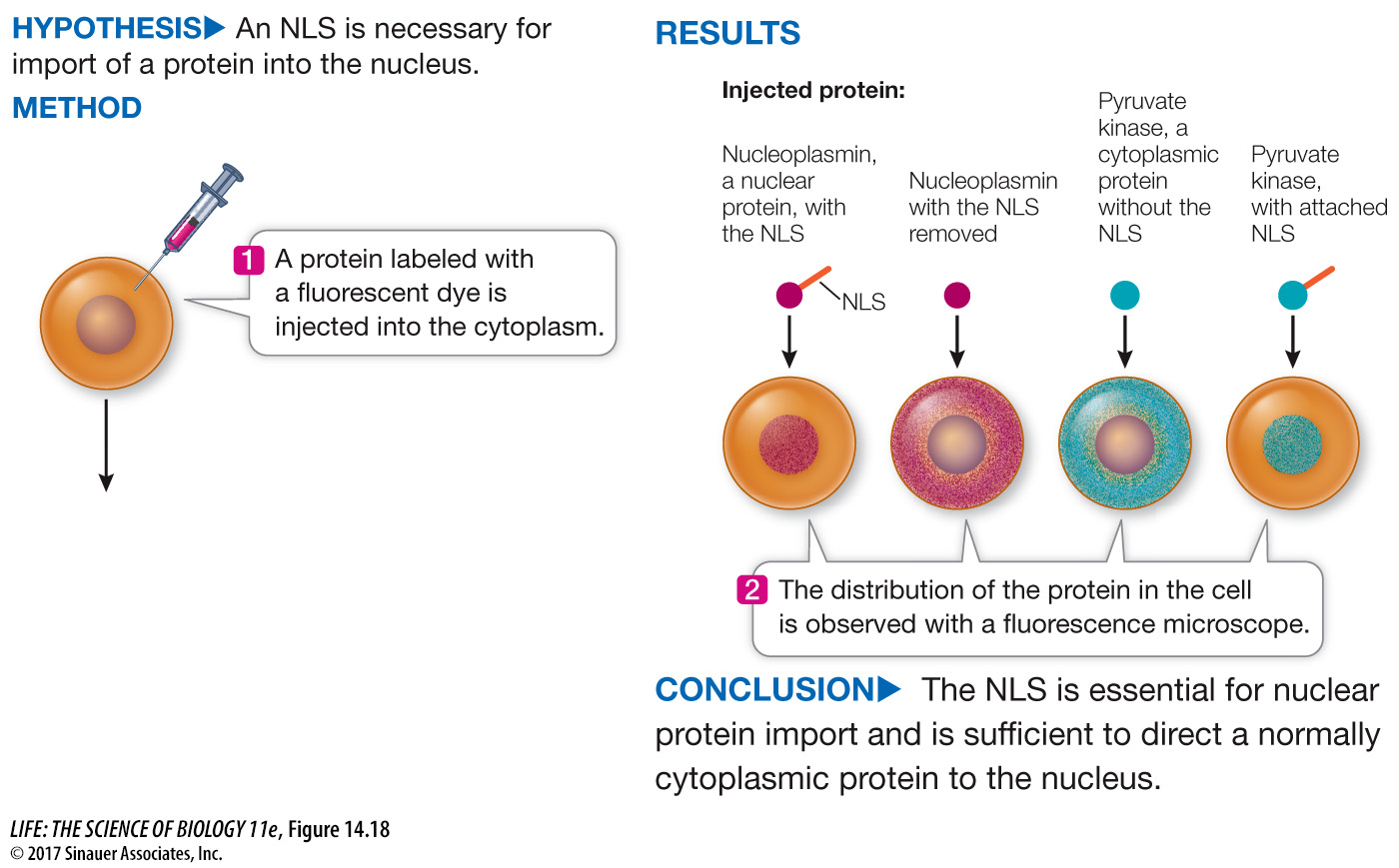
Figure 14.18 Testing the Signal
Original Paper: Dingwall, C. et al. 1988. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-
A. Richardson and his colleagues performed a series of experiments to test whether the nuclear localization signal (NLS) is all that is needed to direct a protein to the nucleus.
If a polypeptide carries a signal of about 20 hydrophobic amino acids at its N terminus, it will be directed to the rough endoplasmic reticulum (RER) for further processing (see Figure 14.17). Note that this is not a specific sequence of amino acids, just a generally hydrophobic sequence at the N terminus that is first translated. Translation will pause, and the ribosome will bind to a receptor at the RER membrane. Once the polypeptide–
The importance of signals is shown by Inclusion-