Cleavage of DNA by restriction enzymes can be used to rapidly detect mutations
All organisms, including bacteria, must have ways of dealing with their enemies. As we saw in Key Concept 13.1, bacteria can be attacked by viruses called bacteriophages. These viruses inject their genetic material into the host cell and turn it into a virus-
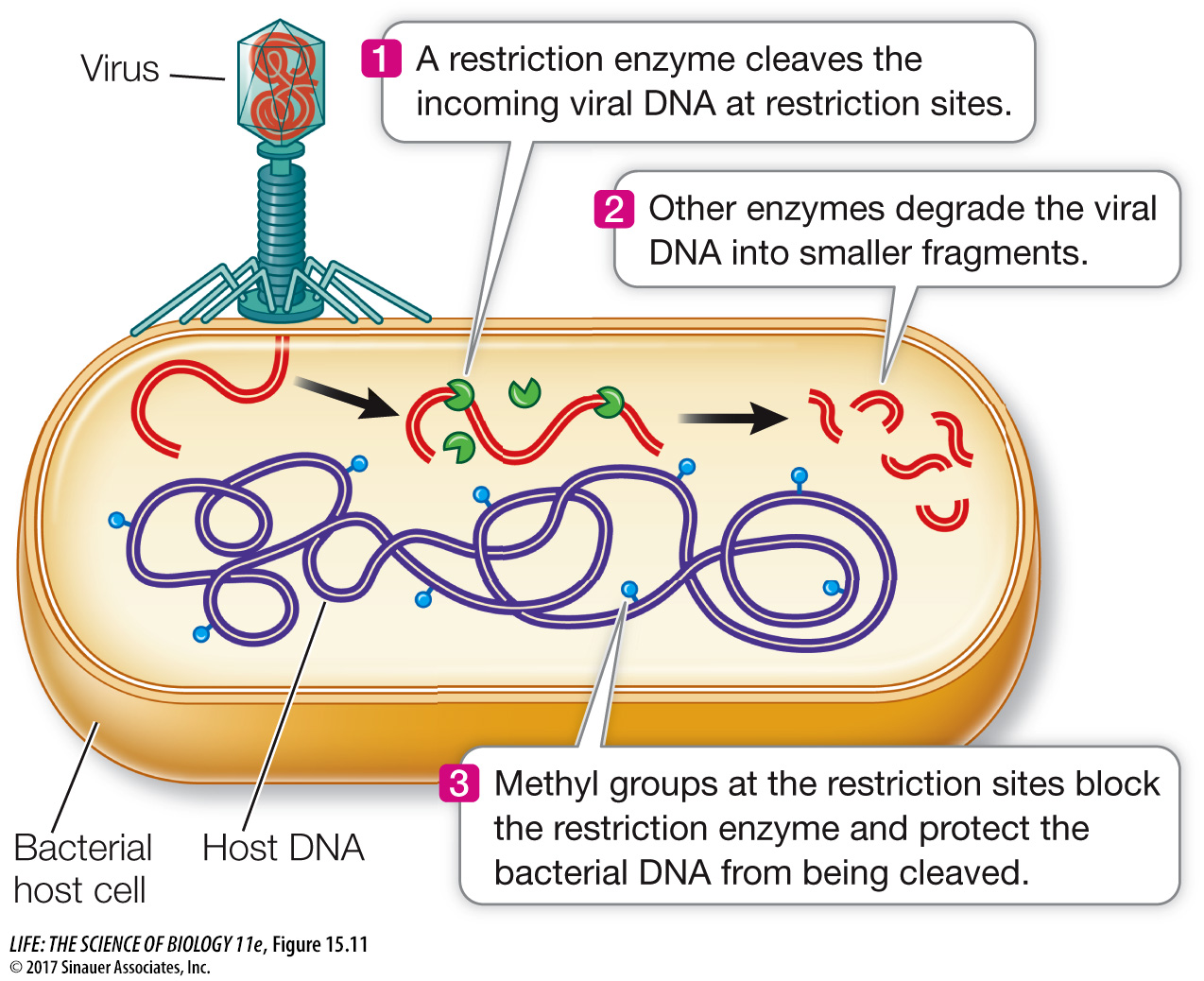
There are many such restriction enzymes, each of which cleaves DNA at a specific sequence of bases called a recognition sequence or a restriction site. Most recognition sequences are four to six base pairs long. Because each sequence of bases has a unique structure (see Key Concept 13.2), it can be specifically recognized by a particular restriction enzyme. Cells protect themselves from being digested by their own enzymes by modifying their DNA, often with methyl groups, to prevent binding by the restriction enzymes.
Restriction enzymes can be isolated from the cells that make them and used as biochemical reagents in the laboratory to give information about the nucleotide sequences of DNA molecules from other organisms. If DNA from any organism is incubated in a test tube with a restriction enzyme (along with buffers and salts that help the enzyme function), that DNA will be cut wherever the restriction site occurs. A specific sequence of bases defines each restriction site. For example, the enzyme EcoRI (named after its source strain of the bacterium E. coli) cuts DNA only where it encounters the following paired sequence in the DNA double helix:
324
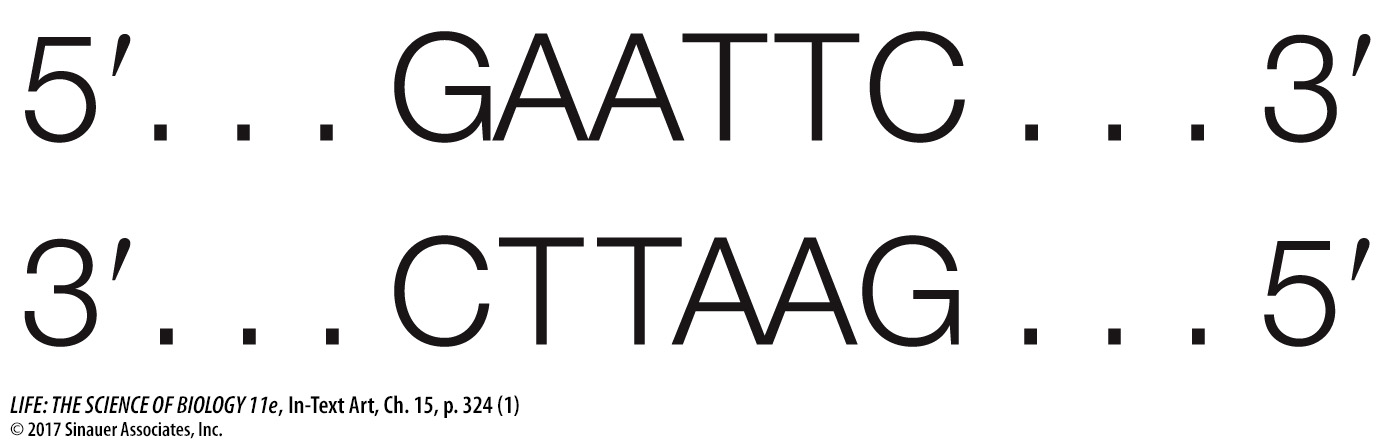
Note that this sequence is palindromic, like the word “mom.” This means that both strands have the same sequence when they are read from their 5′ (or their 3′) ends. The EcoRI enzyme has two identical active sites on its two subunits, which cleave the two strands simultaneously between the G and the A of each strand:
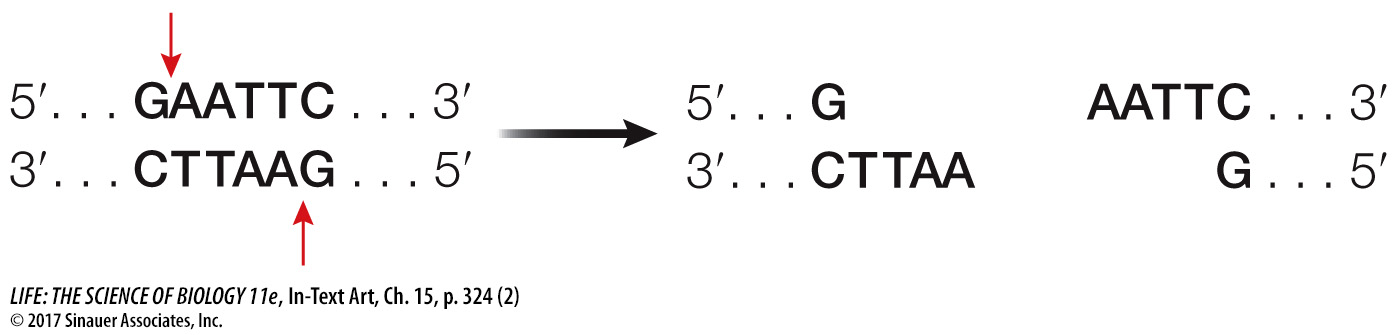
The EcoRI recognition sequence occurs, on average, about once in every 4,000 base pairs in a typical prokaryotic genome, or about once per four prokaryotic genes. So EcoRI can chop a large piece of DNA into smaller pieces containing, on average, just a few genes. Using EcoRI in the laboratory to cut small genomes, such as those of viruses that have tens of thousands of base pairs, may result in just a few fragments. For a huge eukaryotic chromosome with tens of millions of base pairs, a very large number of fragments will be created.
Of course, “on average” does not mean that the enzyme cuts all stretches of DNA at regular intervals. For example, the EcoRI recognition sequence does not occur even once in the 40,000 base pairs of the T7 bacteriophage genome—