DNA fingerprinting combines PCR with restriction analysis and electrophoresis
The methods we have just described are used in DNA fingerprinting, which identifies individuals based on differences in their DNA sequences. DNA fingerprinting works best with sequences that are highly polymorphic—
325
Single nucleotide polymorphisms (SNPs; pronounced “snips”) are inherited variations involving a single nucleotide base—
they are point mutations. These polymorphisms have been mapped for many organisms. If one parent is homozygous for the base A at a certain point in the genome, and the other parent is homozygous for a G at that point, the offspring will be heterozygous: one chromosome will have A at that point and the other will have G. If a SNP occurs in a restriction enzyme recognition site, such that one variant is recognized by the enzyme and the other isn’t, then individuals can be distinguished from one another very easily using the *polymerase chain reaction (PCR). A fragment containing the polymorphic sequence is amplified by PCR from samples of total DNA isolated from each individual. The fragments are then cut with the restriction enzyme and analyzed by gel electrophoresis. *connect the concepts As described in Key Concept 13.5, PCR is a technique used to make multiple copies of a DNA sequence—
that is, to amplify it. The process essentially automates DNA replication; over many cycles the amount of DNA can be increased exponentially. Activity 15.3 Allele-
Specific Cleavage www.life11e.com/
ac15.3 Short tandem repeats (STRs) are short, repetitive DNA sequences that occur side by side on the chromosomes, usually in the noncoding regions. These repeat patterns, which contain one to five base pairs, are also inherited. For example, at a particular locus on chromosome 15 there may be an STR of “AGG.” An individual may inherit an allele with six copies of the repeat (AGGAGGAGGAGGAGGAGG) from her mother and an allele with two copies (AGGAGG) from her father. Again, PCR is used to amplify DNA fragments containing these repeated sequences, and then the amplified fragments, which have different sizes because of the different lengths of the repeats, are distinguished by gel electrophoresis (Figure 15.13A).
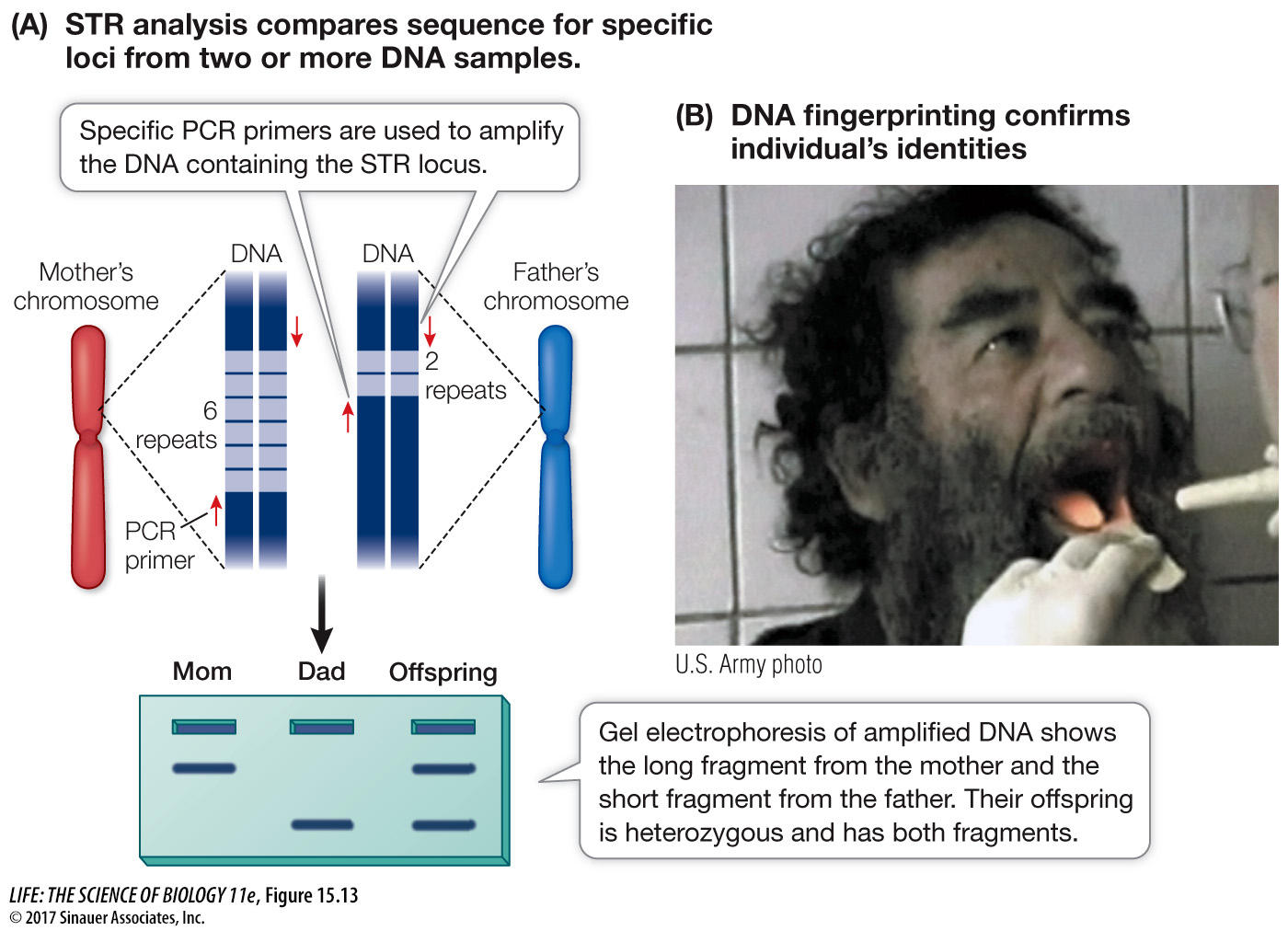 Figure 15.13 DNA Fingerprinting with Short Tandem Repeats (A) A particular STR locus can be analyzed to determine the number of repeat sequences that were inherited by an individual from each parent. The two alleles can be identified in an electrophoresis gel on the basis of their sizes. When several STR loci are analyzed, the pattern can constitute a definitive identification of an individual. (B) When the dictator Saddam Hussein was captured in Iraq, a sample of his cheek epithelial cells was taken for DNA fingerprinting. A comparison with DNA fingerprints of relatives provided military scientists with evidence that the man in question was indeed Saddam Hussein.
Figure 15.13 DNA Fingerprinting with Short Tandem Repeats (A) A particular STR locus can be analyzed to determine the number of repeat sequences that were inherited by an individual from each parent. The two alleles can be identified in an electrophoresis gel on the basis of their sizes. When several STR loci are analyzed, the pattern can constitute a definitive identification of an individual. (B) When the dictator Saddam Hussein was captured in Iraq, a sample of his cheek epithelial cells was taken for DNA fingerprinting. A comparison with DNA fingerprints of relatives provided military scientists with evidence that the man in question was indeed Saddam Hussein.Question
Q: Should everyone have their DNA fingerprinted at birth to create a “genetic ID” for use in screening for genetic diseases? What would be the advantages and disadvantages of this?
The advantage of a genetic ID would be to predict future propensities for diseases and possibly act to prevent them, and to identify a person in an accident, war, or crime (this is already done with soldiers and people in federal prisons). A disadvantage would be that in the wrong hands there could be an invasion of privacy. Genetic markers would need to be used with caution as they are not necessarily predictive; environmental factors often play a role in disease as well. While a federal law in the United States prohibits using genetic data to discriminate against people applying for insurance and employment, the concern remains that genetic data may nevertheless influence decision makers in these and other arenas.
The method of DNA fingerprinting used most commonly today hydrolyzes STR analysis. The Federal Bureau of Investigation in the United States uses 13 STR loci in its Combined DNA Index System (CODIS) database (Table 15.3). An analysis of these loci in your DNA would reveal your particular DNA fingerprint. Looking at Table 15.3, you might inherit:
From your mother: allele 72 from chromosome 4; allele 23 from chromosome 7; allele 14 from chromosome 11; and allele 12 from chromosome 18
From your father: allele 56 from chromosome 4; allele 22 from chromosome 7; allele 16 from chromosome 11; and allele 12 from chromosome 18
| Human chromosome | Locus name | Repeated sequence | Number of alleles |
|---|---|---|---|
| 4 | FGA | CTTT | 80 |
| 7 | D7S820 | GATA | 30 |
| 11 | TH01 | TCAT | 20 |
| 18 | S18S51 | AGAA | 51 |
326
Note that in this case you are heterozygous for the alleles on three of the chromosomes and homozygous for the allele on chromosome 18. With all the alleles and 13 loci, the probability of two people sharing the same alleles is very small. So a DNA sample from a crime scene can be used to determine whether a particular suspect left that sample at the scene.
As we have just shown, DNA fingerprinting can be used to help prove the innocence or guilt of a suspect, but it can also be used to identify individuals who are related to one another. On May 2, 2011, Osama bin Laden was killed by U.S. soldiers at his home in Pakistan. He was identified at the scene by comparison with photographs, a wife who pointed him out, and instant analysis using a digital camera with facial recognition software. In addition, DNA fingerprinting was used. Bin Laden’s son Khalid was also killed in the raid, and a sister had previously died in the U.S. Analyses of their DNA along with that of Osama indicated that the three shared many polymorphisms and were highly likely to be closely related. The same methods were also used to identify Saddam Hussein, who was captured in 2003 and later executed in Iraq (Figure 15.13B).
DNA analysis with genetic markers such as SNPs and STRs has applications throughout all areas of biological research. For example, these markers are used to analyze the organization of genomes, to identify species or individuals within species, to compare species or organisms to see how closely related they are, and to analyze particular genes and the phenotypes associated with them. In the remainder of this chapter we will focus on the use of these markers and other technologies that are used to study and treat genetic diseases.