Genetic markers can be used to find disease-
327
Identifying a mutant gene requires finding a marker that is closely linked to the gene of interest, a process called linkage analysis. The reference points for gene isolation are genetic markers. Genetic markers such as STRs and SNPs can be used as landmarks to find a gene of interest, if the gene also has multiple alleles (for example, normal and disease-
As you have seen, SNPs and STRs are widespread in eukaryotic genomes. There is roughly one SNP for every 1,330 base pairs in the human genome, and any regions of the genome also contain repetitive DNA sequences such as those found in STRs. SNPs and STRs can be analyzed using the PCR techniques mentioned above. SNPs can also be detected using sophisticated chemical methods such as mass spectrometry.
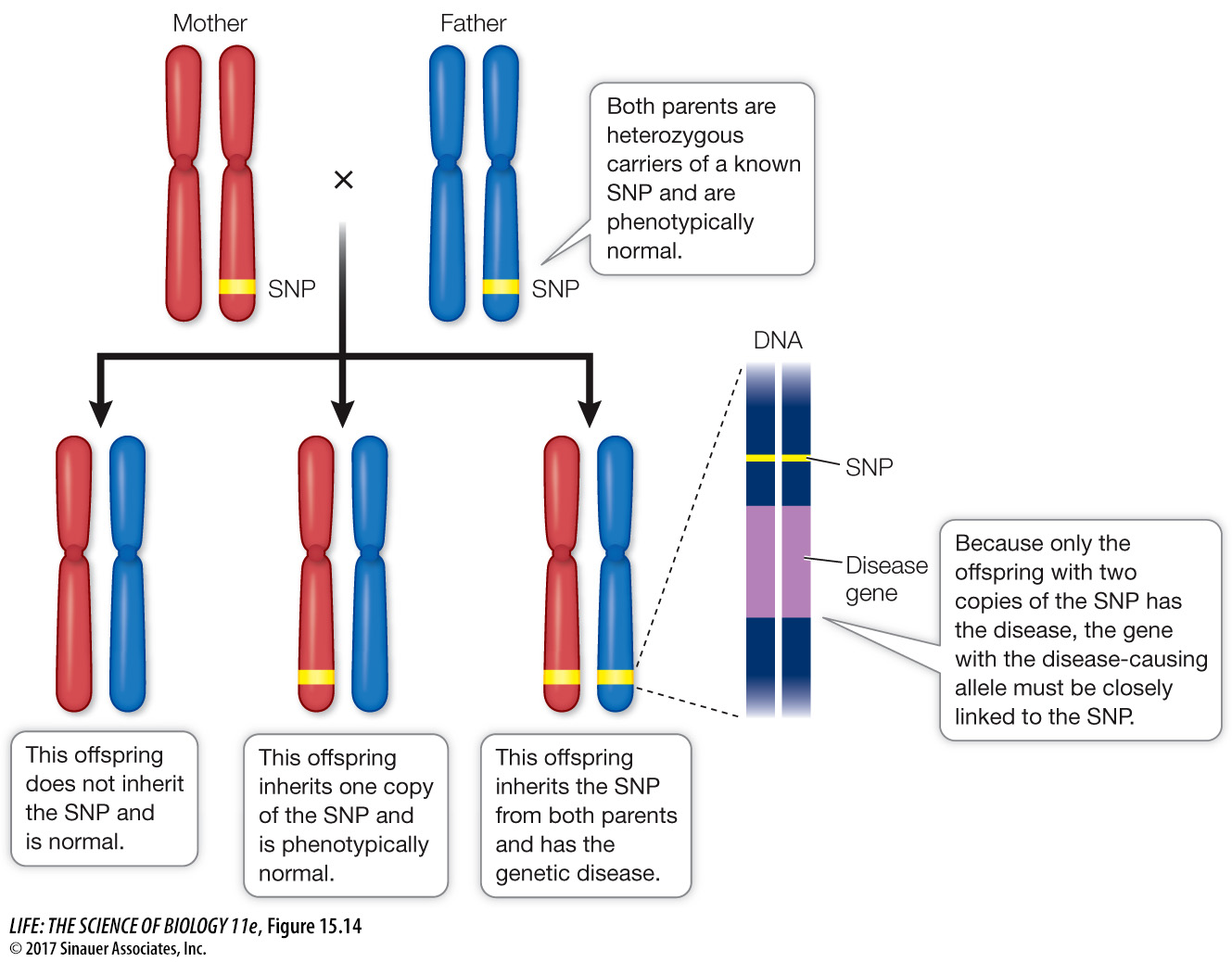
To narrow down the location of a gene, a scientist must find a genetic marker that is always inherited with the gene. To do this, family medical histories are taken and pedigrees are constructed. If a genetic marker and a genetic disease are inherited together in many families, then they must be near each other on the same chromosome (Figure 15.14). This situation recalls the conclusion reached in the classic studies of inheritance undertaken by Thomas Hunt Morgan and discussed in Key Concept 12.4: two genes do not always assort independently. Genes that are “linked” on the same chromosome will sometimes be inherited together—
Question
Q: How is DNA linkage analysis similar to chromosome linkage analysis in genetics? How is it different?
Chromosome linkage analysis involves actual crosses and is done by analyzing the results of recombination between alleles. DNA linkage analysis is done on single chromosomes by retrospective analysis of mating. Chromosome linkage analysis examines phenotypes, whereas DNA linkage analysis examines genotypes (DNA).
Linkage narrows down the location of the gene to a few hundred thousand base pairs. Once a linked DNA region is identified, many methods are available to identify the actual gene responsible for a genetic disease. The complete sequence of the region can be searched for candidate genes, using information available from databases of genome sequences. With luck a scientist can make an educated guess, based on biochemical or physiological information about the disease, along with information about the functions of candidate genes, as to which gene is responsible for the disease. The identification of DNA polymorphisms within candidate genes that correlate with the presence or absence of disease can also help narrow down the search. A variety of techniques, such as analyzing mRNA levels of candidate genes in diseased and healthy individuals, are used to confirm that the correct gene has been identified.
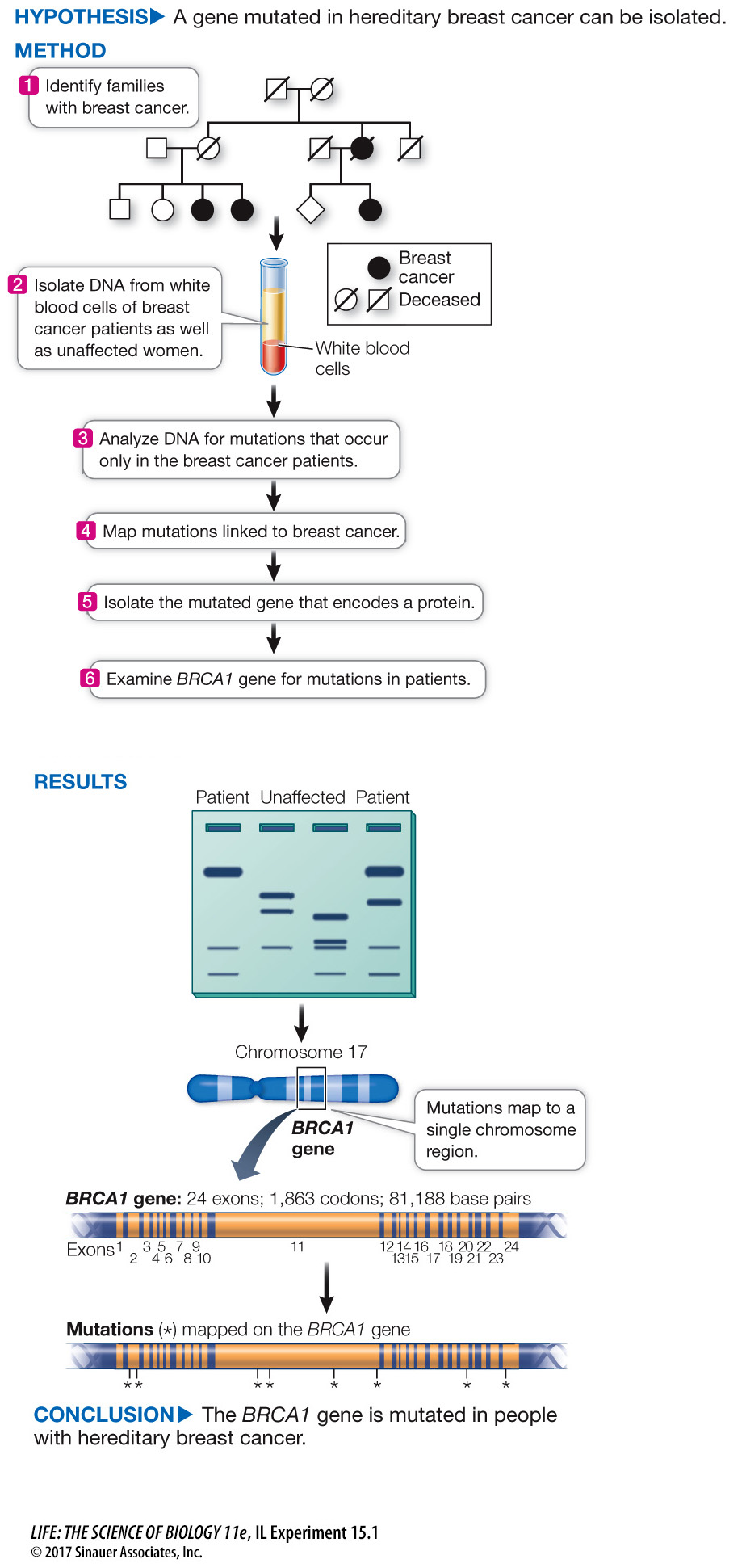
The isolation of the BRCA1 gene that is involved in breast cancer (described in the chapter opening) offers a good illustration of molecular techniques used to identify genes associated with disease, and is described in Investigating Life: How Was the BRCA1 Gene Identified? Analysis of the BRCA1 mutations came after Marie-
328
investigating life
How Was the BRCA1 Gene Identified?
experiment
Original Paper: Miki, Y. et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–
DNA linkage analysis of families led to the isolation of BRCA1, a gene that can lead to breast cancer.
work with the data
About one in ten cases of breast cancer is hereditary. That is, the cancer arises from a germ line mutation (meaning it occurs in all cells of the body) that is expressed in breast cells as cancer. The other nine in ten cases come about from somatic mutations in breast cells. The identification of a gene involved in hereditary breast cancer was made by DNA analysis. First, the location of the gene was narrowed down by analysis of SNPs in families where several women had breast cancer (see the experiment). Then a team led by Mark Skolnick at the University of Utah and the company Myriad Genetics sequenced the DNA in the candidate region and looked for a sequence that had a promoter, transcription terminator, start and stop codons, and sequences at the ends of introns—
QUESTIONS
Question 1
Hereditary (as opposed to somatic) breast cancer is often suspected when the cancer develops relatively early in life. Hereditary cancers tend to arise earlier because they involve a germ line mutation present in all cells at birth, whereas a cancer due to a spontaneous somatic mutation in the DNA of a breast cell can happen any time (statistically, later). Skolnick’s team examined the timing of breast cancer cases in patients who had different mutations in BRCA1. The results are shown in Table A. What conclusions can you draw?
| Family | Total cases | Cases before age 50 |
|---|---|---|
| A | 31 | 20 |
| B | 22 | 14 |
| C | 10 | 7 |
In the three families, breast cancer occurred in two-
Question 2
DNA sequencing evaluated BRCA1 in the cancer patients and their families, as well as in controls who did not have breast cancer themselves or in their family. The results are shown in Table B. Were the mutations in BRCA1 present only in the breast cancer patients? What do you think the effects of the mutations were in each of the three families?
| Family | Codon number in BRCA1 |
Change in patients | Change in controls? |
|---|---|---|---|
| A | 1313 | C → T | None (0/170) |
| B | 1775 | T → G | None (0/120) |
| C | 24 | deletion 11 bp | None (0/180) |
All three mutations were present only in the breast cancer patients. In Families A and B, the two point mutations may have caused codon mutations and a different amino acid in each case in the BRAC1 protein, causing it to be nonfunctional. In Family C, the deletion was 11 base pairs, which resulted in a frame-
Question 3
Human tissues from people without breast cancer were examined by nucleic acid hybridization (see Figure 14.6) for the level of mRNA for BRCA1. The results are shown in Table C. What can you conclude about the kinds of cancer that might be caused by mutations in BRCA1?
| Tissue | mRNA detected? |
|---|---|
| Breast | Yes |
| Colon | No |
| Intestine | No |
| Ovary | Yes |
| Prostate | No |
| Spleen | No |
| Testis | No |
| Thymus | Yes |
BRCA1 is active in breast, ovary, and thymus tissues. So mutations would also be expressed there. Since BRCA1 is involved in DNA repair, all three of these tissues would experience poor repair, leading to additional mutations that can cause cancer.
A similar work with the data exercise may be assigned in LaunchPad.