Epigenetic changes can be induced by the environment
Female honey bees all have the same genetic makeup. When they are in the immature stage called a larva, however, one female in the hive eats a protein-
Although they are reversible, many epigenetic changes such as DNA methylation and histone modification can permanently alter gene expression patterns in a cell. In a germ line cell that forms gametes, epigenetic changes can be passed on to the next generation. But what determines these epigenetic changes? A clue comes from a recent study of monozygotic (identical) twins. Monozygotic twins come from a single fertilized egg that divides to produce two separate cells; each of these goes on to develop a separate individual. Monozygotic twins thus have identical genomes. But are they identical in their epigenomes? A comparison of DNA in hundreds of such twin pairs shows that in tissues of 3-
351
investigating life
Gene Expression and Behavior
experiment
Original Paper: Kucharski, R., J. Maleszka, S. Foret and R. Maleszka. 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319: 1817–
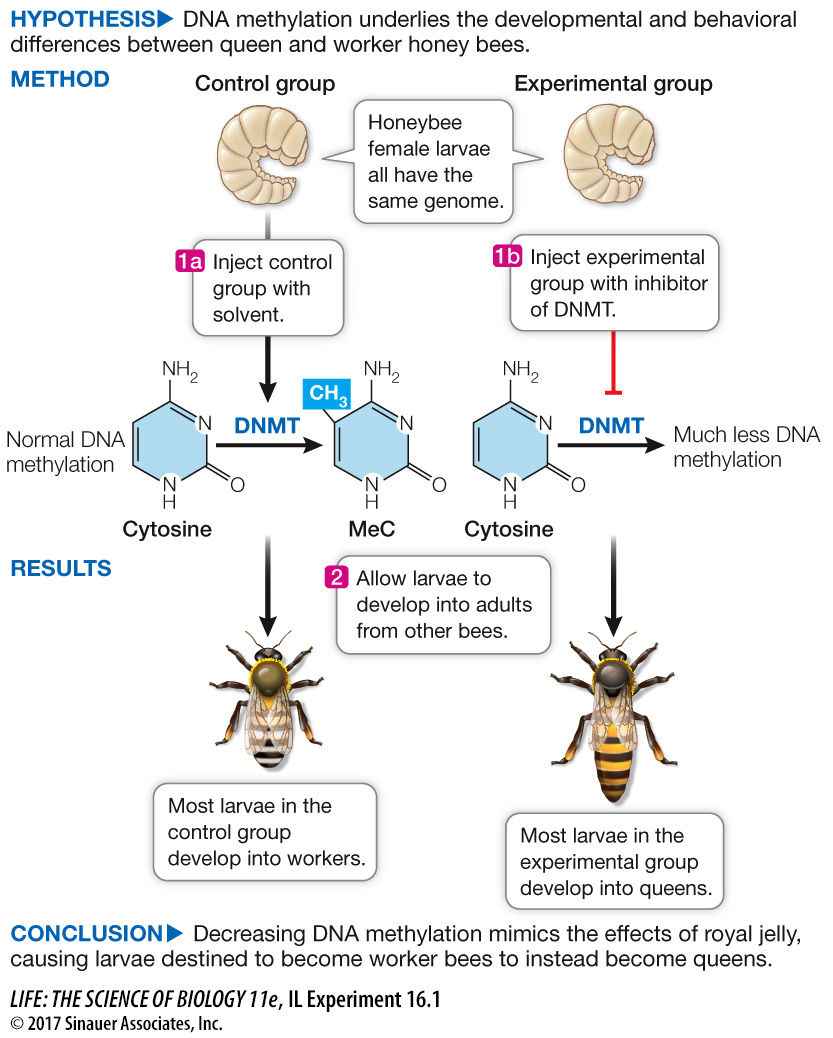
Most female honey bee larvae grow up to be workers; just one eats royal jelly and becomes the queen. To understand differences in gene expression in queen bees versus their sisters with identical genomes, Ryszard Maleszka and colleagues at Australian National University developed an experiment to test for possible epigenetic effects.
The experiment involved injecting female honey bee larvae with a substance that inhibits expression of the enzyme DNA cytosine-
work with the data
QUESTIONS
Question 1
After injection with either an inhibitor of DNMT or a control, the level of DNMT mRNA was measured in the heads of the larvae and compared with the level of a control mRNA that is always expressed at a high level. The results are shown in Table A.
| Time (h) | DNMT mRNA level compared to control (%) |
|---|---|
| 23 | 105 |
| 48 | 41 |
Why was mRNA measured in the heads of the larvae?
What can you conclude about the effectiveness of the inhibition of DNMT expression?
mRNA was measured in the heads because gene expression in the brain determines whether a honey bee will be a worker or a queen.
The level of inhibition of DNMT mRNA was about 60 percent after 48 h. This is good but not perfect inhibition. So some DNMT mRNA probably remained.
Question 2
The DNA of a gene that is normally expressed in the brain (head) of the larvae was sequenced and the percentage of 5–
| Condition | Percent 5– |
|---|---|
| Control | 79 |
| DNMT expression inhibited | 63 |
The reduction of DNA methylation was about 20 percent. So some 5′-methylcytosine remained.
Question 3
Larvae that had been injected with the inhibitor for DNMT gene expression or a control were allowed to develop into adults. The phenotypes of the adults were evaluated, and the data are shown in Table C. What do the data show about the effect of the inhibition of DNA methylation? Does the extent of DNA methylation strictly correlate with the extent of phenotypic change?
| Condition | Number of workers | Number of queens |
|---|---|---|
| Control | 238 | 73 |
| Inhibited | 74 | 188 |
In the controls, about 23 percent of the larvae developed into queens. But in the larvae that had reduced DNA methylation, about 72 percent developed into queens. This is a remarkable shift given that DNMT mRNA and DNA methylation were reduced but not as much. There must be a threshold for methylation, above which gene expression is affected.
A similar work with the data exercise may be assigned in LaunchPad.
352
Media Clip 16.1 The Surprising Epigenetics of Identical Twins
www.Life11e.com/