Operons are units of transcriptional regulation in prokaryotes
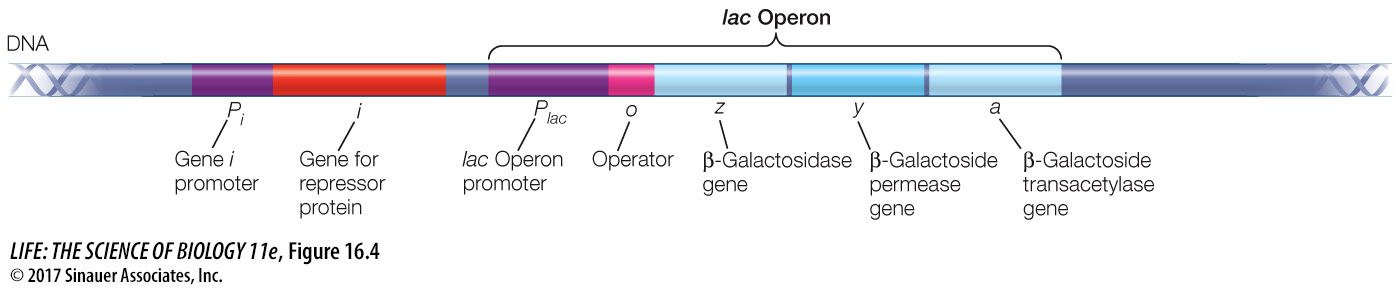
The genes in E. coli that encode the three enzymes for using lactose are structural genes; structural genes specify the primary structures (the amino acid sequences) of protein molecules that act as enzymes or cytoskeletal proteins. The three genes are adjacent to one another on the E. coli chromosome. This arrangement is no coincidence: the genes share a single promoter, and their DNA is transcribed into a single, continuous molecule of mRNA. Because this particular mRNA governs the synthesis of all three lactose-
339
A cluster of genes with a single promoter is called an operon, and the operon that encodes the three lactose-
There are numerous mechanisms to control the transcription of operons; we will describe three examples:
An inducible operon regulated by a repressor protein
A repressible operon regulated by a repressor protein
An operon regulated by an activator protein