Some sequences of DNA can move about the genome
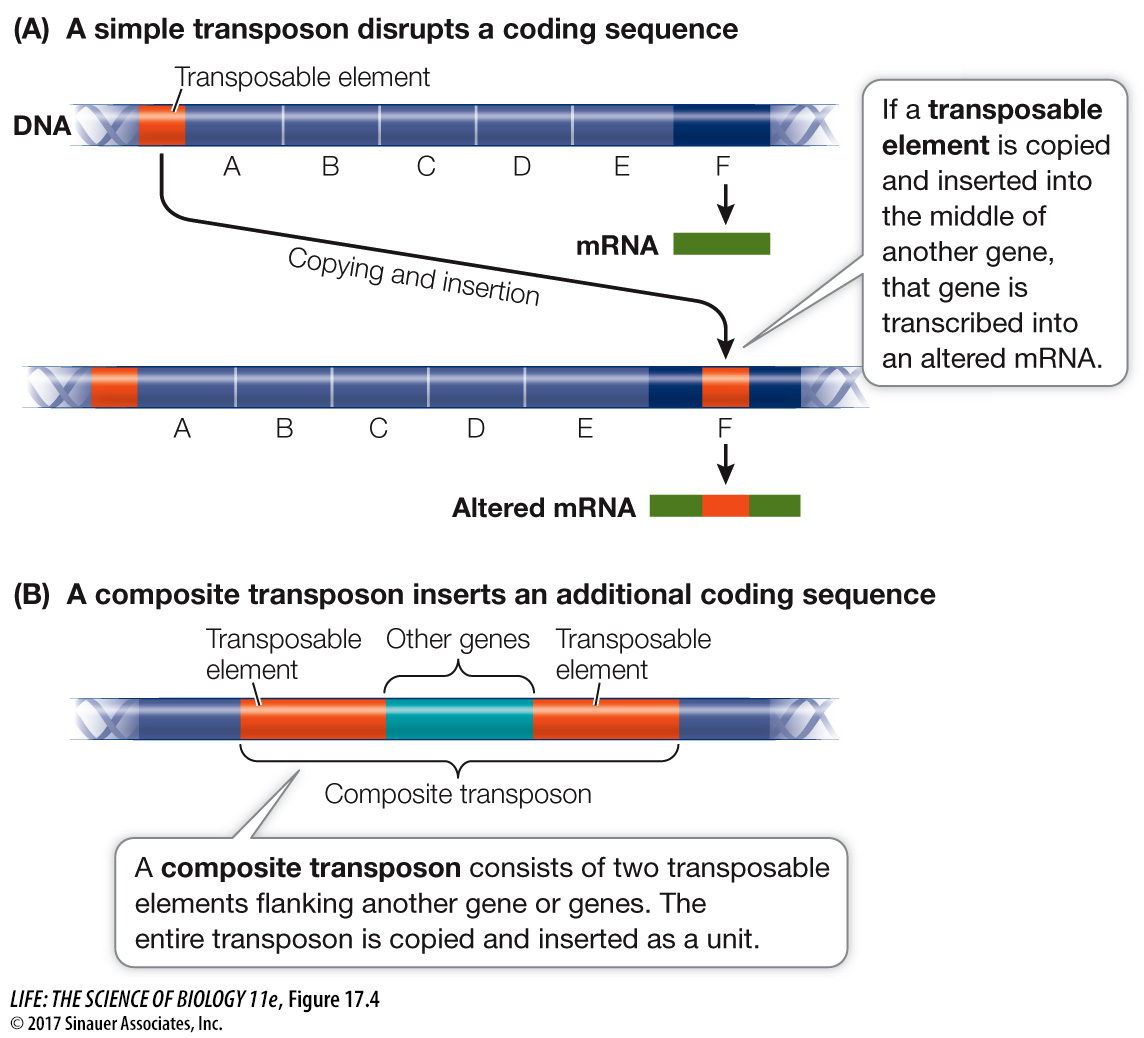
As we mentioned in Key Concept 15.1, transposable elements (or transposons) are segments of DNA that can move from place to place in the genome. Genome sequencing has allowed scientists to study these elements more broadly, and they are now known to be widespread in both prokaryotes and eukaryotes. Prokaryotic transposable elements are often short sequences of 1,000 to 2,000 bp, and they can be found both in chromosomes and in plasmids. A transposable element might be at one location in the genome of one E. coli strain, and at a different location in another strain. The insertion of this movable DNA sequence from elsewhere in the genome into the middle of a protein-
Question
Q: One hypothesis for the origin of some transposons is through a retroviral infection. Based on what you know of the life cycle of a retrovirus (see Key Concept 16.3), how might this occur?
A retrovirus infects a cell and makes a cDNA copy of its genome. The cDNA is inserted into the host cell by the action of viral integrase. The cDNA is carried along with the host chromosome as the cell divides. Through recombination, the cDNA adds a gene for excision. It also adds adjacent genes and is able to move about the genome.
As you saw in Chapter 15, there are several ways for transposable elements to move around the genome. For example, a transposable element may be replicated, and then the copy can be inserted into another site in the genome (the “copy and paste” mode). Or the element might splice out of one location and move to another location (“cut and paste”). Transposable elements usually carry genes for enzymes such as transposases, which catalyze the reactions needed for transposition. Often the elements are flanked by inverted repeat DNA sequences that are recognized by these enzymes.