Eukaryotic genomes contain repetitive sequences
Eukaryotic genomes contain numerous repetitive DNA sequences that do not code for polypeptides. These are typically not in protein-
Highly repetitive sequences are short (less than 100 bp) sequences that are repeated thousands of times in tandem (side-
372
*connect the concepts Key Concept 15.3 described how STRs can be used in the identification of individuals (DNA fingerprinting).
Moderately repetitive sequences are repeated 10 to 1,000 times in the eukaryotic genomes. These sequences include the genes that are transcribed to produce tRNAs and rRNAs, which are used in protein synthesis. The cell makes tRNAs and rRNAs constantly, but even at the maximum rate of transcription, single copies of the tRNA and rRNA genes would be inadequate to supply the large amounts of these molecules needed by most cells. Thus the genome has multiple copies of these genes.
In mammals, four different rRNA molecules make up the ribosome: the 18S, 5.8S, 28S, and 5S rRNAs. (The S stands for Svedberg unit, which is a measure of size.) The 18S, 5.8S, and 28S rRNAs are transcribed together as a single precursor RNA molecule (Figure 17.10). As a result of several posttranscriptional steps, the precursor is cut into the final three rRNA products, and the noncoding “spacer” RNA is discarded. The sequence encoding these RNAs is moderately repetitive in humans: a total of 280 copies of the sequence are located in clusters on five different chromosomes.
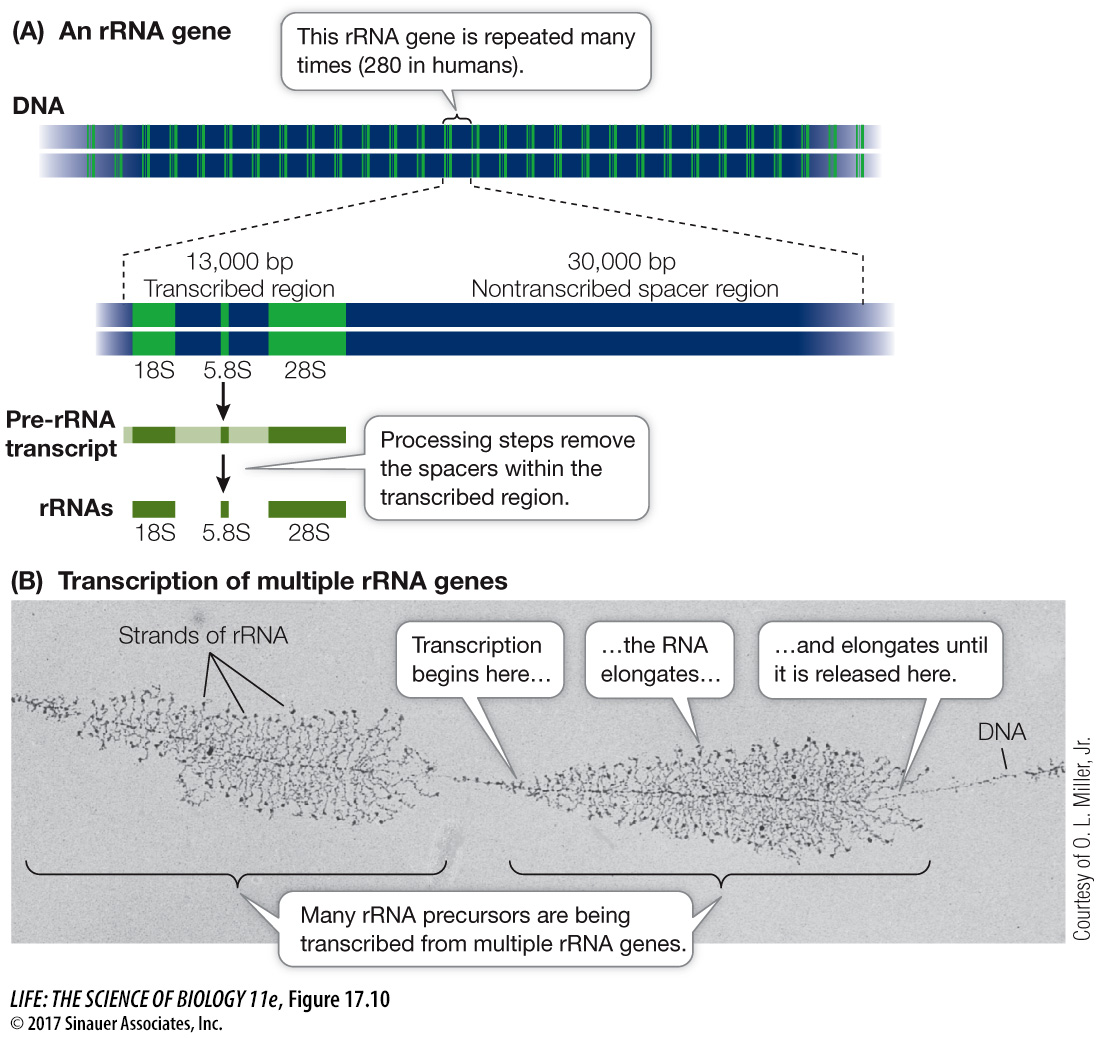
Question
Q: Are there similarities between the process depicted in the photo here and the process of translation via polysome shown in Figure 14.16?
In both processes a protein structure is moving along a nucleic acid. In the polysome, the ribosome moves along mRNA; in rRNA synthesis, RNA polymerase moves along DNA. In both processes a polymer product is made: in the polysome the product is a polypeptide, and in the rRNA the product is RNA. The two processes are similar also in having a “cafeteria” system in which multiple polymers are made at the same time.
Apart from the RNA genes, most moderately repetitive sequences are transposons, which, like the prokaryotic transposons we discussed earlier, can move about in the genome. Transposons make up more than 40 percent of the human genome and about 50 percent of the corn genome, but in many other eukaryotes the percentage is smaller (3–
Table 17.6 summarizes the major types of transposons in eukaryotes. Retrotranspons fall into three groups, based on the types of repetitive sequences they contain: long terminal repeats (LTRs), long interspersed elements (LINEs), and short interspersed elements (SINEs). Retrotransposons move about the genome in a distinctive way: they are transcribed into RNA, which then acts as a template for new DNA. The new DNA becomes inserted at a new location in the genome. This “copy and paste” mechanism results in two copies of the transposon: one at the original location and the other at a new location. A single type of SINE retrotransposon, the 300-
| Category | Transcribed | Translated |
|---|---|---|
| Single- |
||
| Promoters and expression control sequences | No | No |
| Introns | Yes | No |
| Exons | Yes | Yes |
| Moderately repetitive sequences | ||
| rRNA and tRNA genes | Yes | No |
| Transposons | ||
| I. Retrotransposons | ||
| LTRs | Yes | No |
| SINEs | Yes | No |
| LINEs | Yes | Yes |
| II. DNA transposons | Yes | Yes |
| Highly repetitive short sequences | No | No |
Transposons of the fourth type, the DNA transposons, do not use RNA intermediates. Like some prokaryotic transposable elements, they are excised from the original location and become inserted at a new location without being replicated (a “cut and paste” mechanism).
373
What role do these moving sequences play in the cell? The best answer so far seems to be that transposons are simply molecular parasites that can be replicated. The insertion of a transposon at a new location can have important consequences. For example, the insertion of a transposon into the coding region of a gene results in a mutation (see Figure 17.5). This phenomenon accounts for rare forms of several genetic diseases in humans, including hemophilia and muscular dystrophy. If the insertion of a transposon takes place in the germ line, a gamete with a new mutation results. If the insertion takes place in a somatic cell, cancer may result.
Sometimes an adjacent gene can be replicated along with a transposon, resulting in a gene duplication. A transposon can carry a gene, or a part of it, to a new location in the genome, shuffling the genetic material and creating new genes. Clearly, transposition stirs the genetic pot in the eukaryotic genomes and thus contributes to genetic variation.