Human genomics has potential benefits in medicine
Most complex phenotypes are determined not by single genes but by multiple genes interacting with the environment. The single-
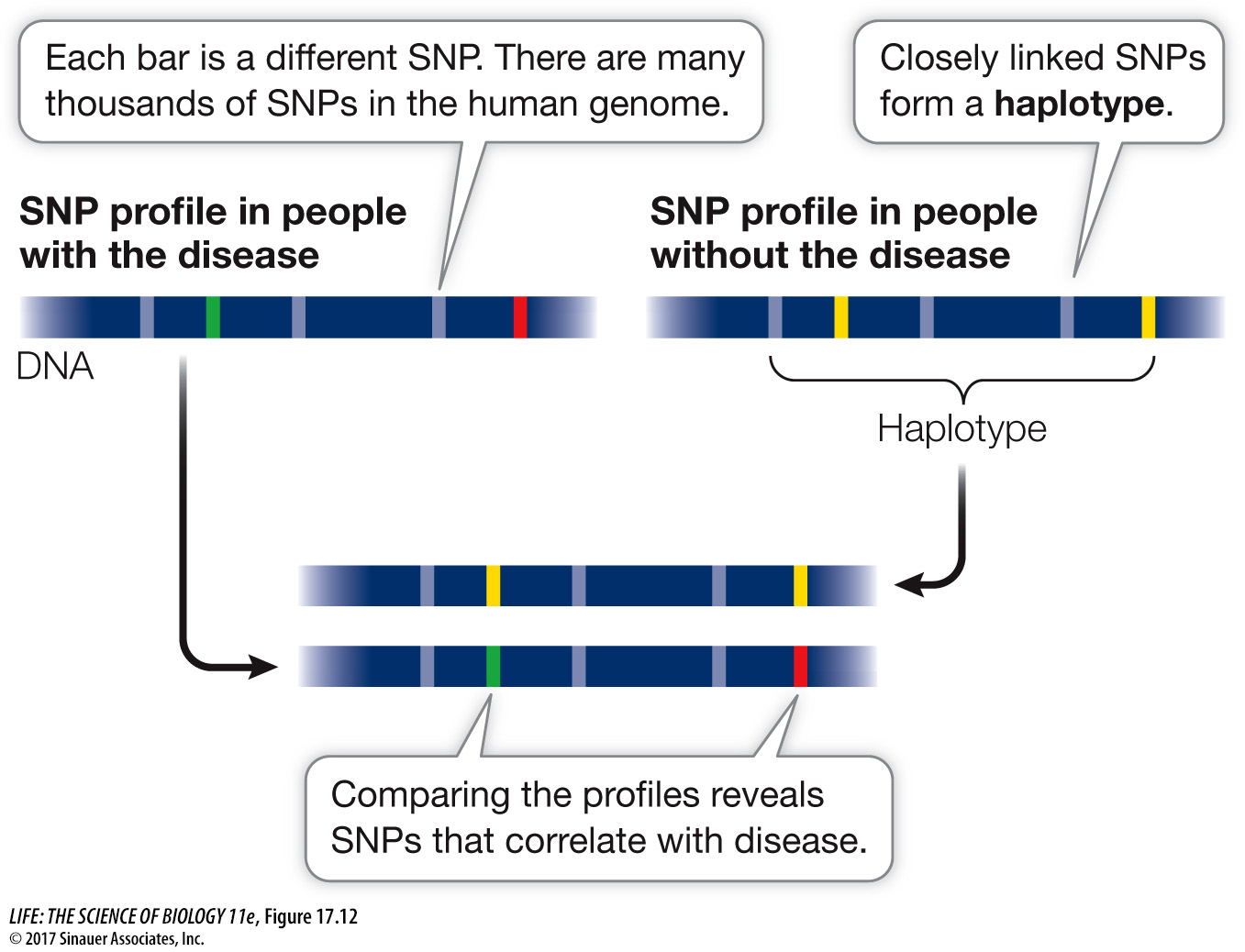
HAPLOTYPE MAPPING The SNPs that differ among individuals are not inherited as independent alleles. Rather, a set of SNPs that are present on a segment of chromosome are usually inherited as a unit. This linked piece of a chromosome is called a haplotype. You can think of the chromosome segment as a sentence, the haplotype as a word, and the SNP as a letter in the word. Analyzing SNPs is faster and less expensive than sequencing whole genomes, so haplotype mapping provides a shortcut for identifying the locations of genes and mutations involved in particular diseases (see Key Concept 15.3). By comparing the haplotypes of individuals with and without a particular genetic disease, the genetic loci associated with the disease can be identified (Figure 17.12).
A microarray of 500,000 SNPs has been used to analyze thousands of people to find out which SNPs are associated with specific diseases. The amount of data is prodigious: 500,000 SNPs, thousands of people, thousands of medical records. With so much natural variation, statistical measures of association between a haplotype and a disease need to be very rigorous.
GENOTYPING TECHNOLOGY AND PERSONAL GENOMICS Association tests like the one described in Figure 17.12 have revealed particular haplotypes or alleles that are associated with modestly increased risks for diseases. For example, a particular SNP on chromosome 10 is associated with a 65% increased risk for adult-
375
Of course, the most comprehensive way to analyze a person’s genome is by actually sequencing it. As the cost of genome sequencing and analysis goes down, SNP testing will be superseded. In 2015 the U.S. government announced a plan to sequence the genomes of 1 million Americans and link the sequences to medical records. This approach is not new, but the large scale of it is. The goal is to relate genome changes to diseases such as cancer and diabetes, and to use these changes for diagnosis and, if they lead to changes in encoded proteins, to therapies targeted at the altered proteins.
PHARMACOGENOMICS Genetic variation can affect how an individual responds to a particular drug. For example, a drug may be chemically modified in the liver to make it more or less active. Consider an enzyme that catalyzes the following reaction:
Active drug → less active drug
A mutation in the gene that encodes this enzyme may make the enzyme less active. For a given dose of the drug, a person with the mutation would have more active drug in the bloodstream than a person without the mutation. So the effective dose of the drug would be lower in the person with the mutation.
Now consider a different case, in which the liver enzyme is needed to make the drug active:
Inactive drug → active drug
A person carrying a mutation in the gene encoding this liver enzyme would not be affected by the drug, since the activating enzyme is not present.
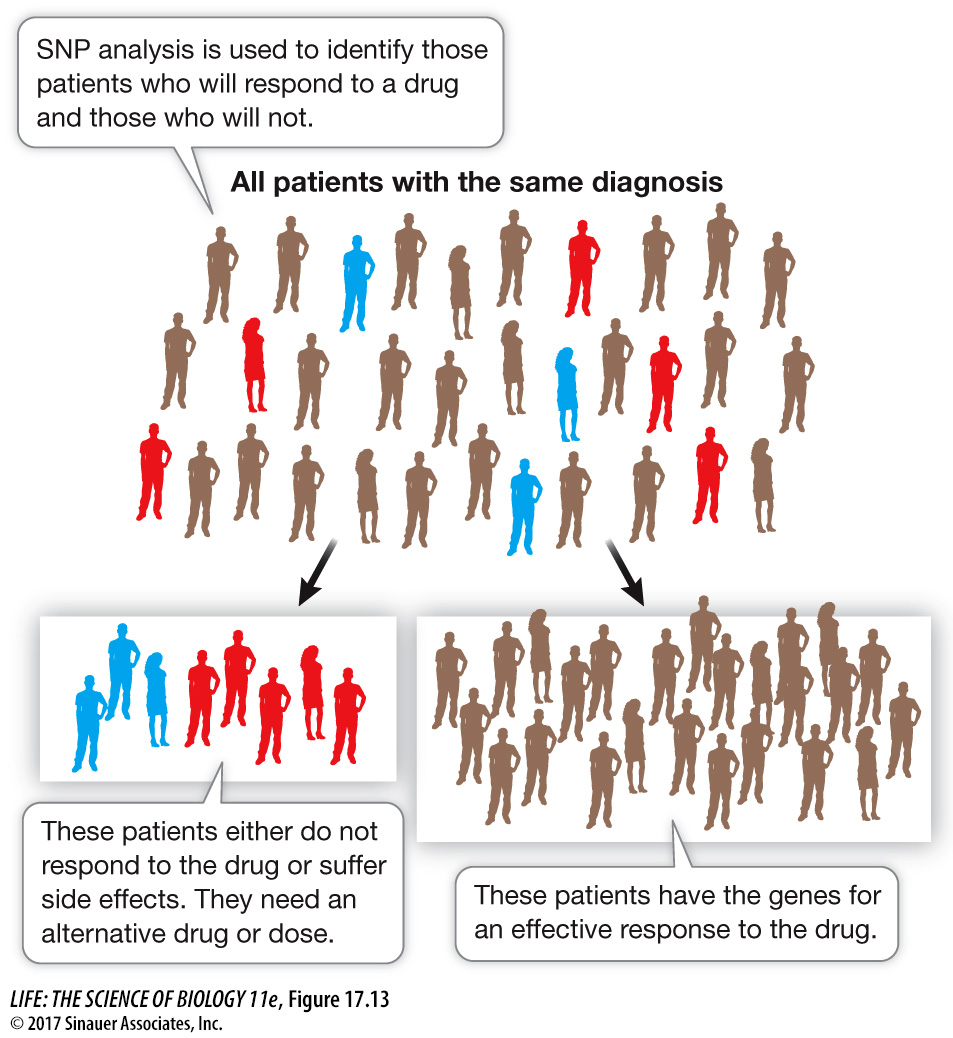
The study of how an individual’s genome affects his or her response to drugs or other outside agents is called pharmacogenomics. Just as it is possible to identify haplotypes or SNPs that are associated with particular disease susceptibilities, it is also possible to identify SNPs that are associated with specific drug responses. This type of analysis makes it possible to predict whether a drug will be effective. The objective is to personalize drug treatment so that a physician can know in advance whether an individual will benefit from a particular drug (Figure 17.13). This approach might also be used to reduce the incidence of adverse drug reactions by identifying individuals who will metabolize a drug slowly, which can lead to a dangerously high level of the drug in the body.