Metagenomics allows us to describe new organisms and ecosystems
If you take a microbiology laboratory course you’ll learn how to identify various prokaryotes on the basis of their growth on particular artificial media. For example, staphylococci are a group of bacteria that infect skin and nasal passages. When grown on a medium called blood agar, they form round, raised colonies. Microorganisms can also be identified by their nutritional requirements or the conditions under which they will grow (for example, aerobic versus anaerobic). Such culture methods have been the mainstay of microbial identification for more than a century and are still useful and important. However, scientists can now use PCR and DNA sequencing to identify microbes without culturing them in the laboratory.
In 1985 Norman Pace, then at Indiana University, came up with the idea of isolating DNA directly from environmental samples. He used PCR to amplify specific rRNA-
Sequencing of DNA from 200 liters of seawater indicated that it contained 5,000 different viruses and 2,000 different bacteria, many of which had not been described previously.
One kilogram of marine sediment contained 1 million different viruses, most of them new.
Water runoff from a mine contained many previously unknown species of prokaryotes thriving in what was previously thought of as an inhospitable environment. Some of these organisms exhibited metabolic pathways that were previously unknown to biologists. These organisms and their capabilities may be useful in cleaning up pollutants from the water.
Gut samples from 124 Europeans revealed that each person harbored at least 160 species of bacteria (making up their gut microflora, or *microbiome). Many of these species were found in all individuals, but the presence of other bacteria varied from person to person. Such variations may be associated with obesity or bowel diseases.
*connect the concepts Learn more about how disruptions of the human microbiome are associated with various autoimmune diseases in Key Concept 25.3.
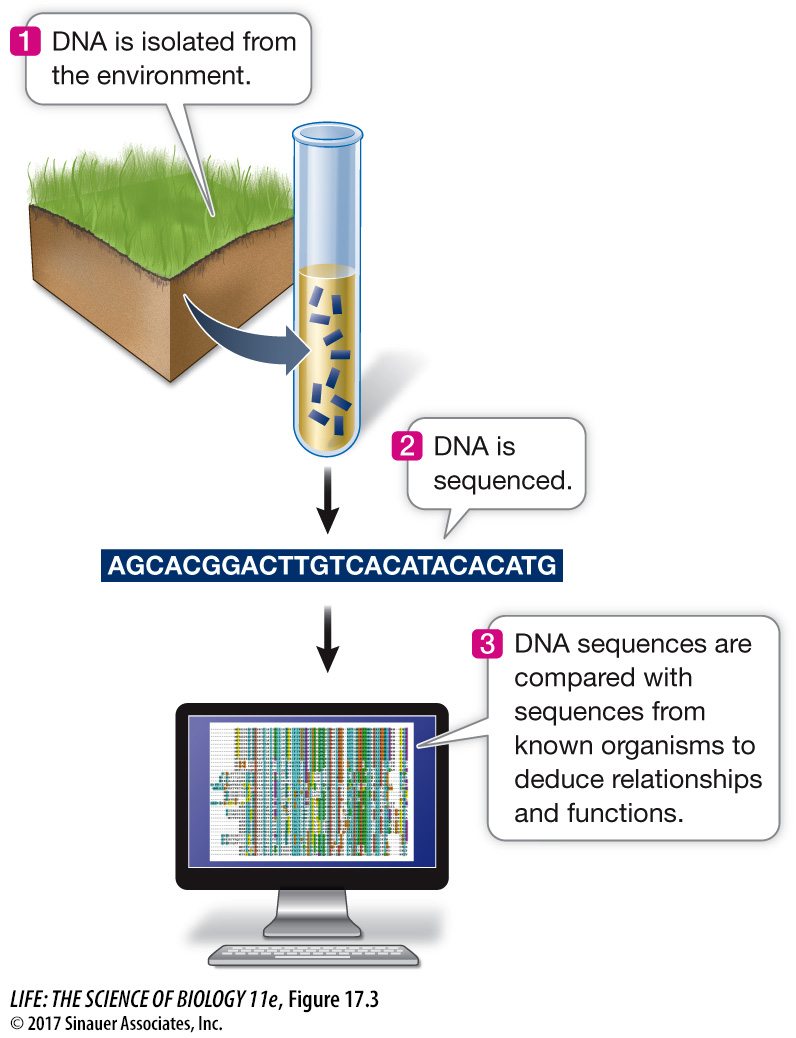
These and other discoveries are truly extraordinary and potentially very important. It is estimated that 90 percent of the microbial world has been invisible to biologists and is only now being revealed by metagenomics. Entirely new ecosystems of bacteria and viruses are being discovered in which, for example, one species produces a molecule that another metabolizes. It is hard to overemphasize the importance of such an increase in our knowledge of the hidden world of microbes. This knowledge will help us understand natural ecological processes, and has the potential to help us find better ways to manage environmental catastrophes such as oil spills, or to remove toxic heavy metals from soil.
366