DNA for cloning can come from a library
A genomic library is a collection of DNA fragments that make up the genome of an organism. Restriction enzyme digestion or other means, such as mechanical shearing, can be used to break chromosomes into smaller pieces. These smaller DNA fragments collectively still constitute a genome (Figure 18.5A), but the information is now in many smaller “volumes.” Any desired fragment can be inserted into a vector, which is then taken up by a host cell. Proliferation of a single such transformed cell produces a colony of cells, each of which harbors many copies of the same fragment of DNA.
research tools
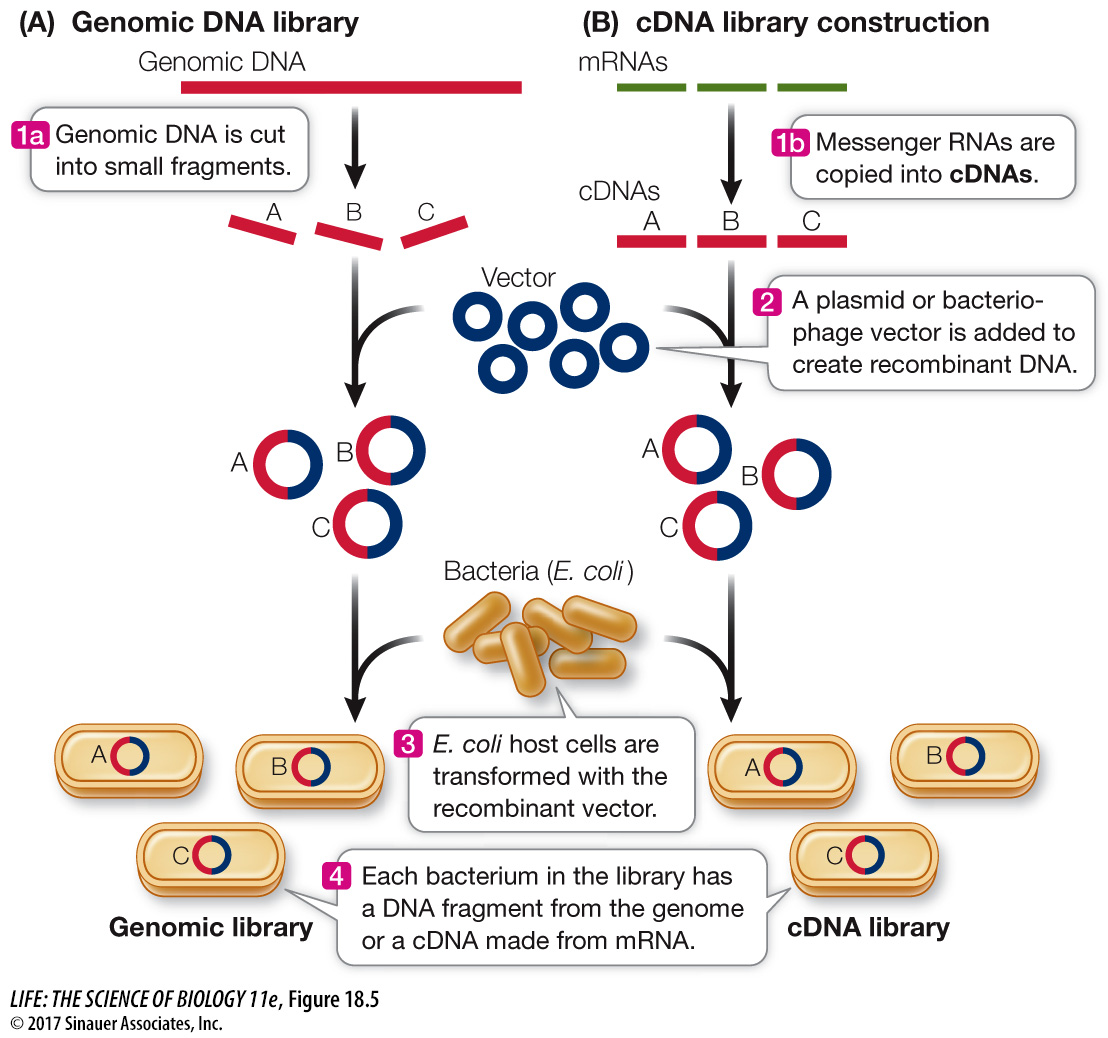
Figure 18.5 Constructing Libraries Intact genomic DNA is too large to be introduced into host cells. (A) A genomic library can be made by breaking the DNA into small fragments, incorporating the fragments into a vector, and then transforming host cells with the recombinant vectors. Each colony of cells contains many copies of a small part of the genome. (B) The many mRNAs in a cell can be copied into cDNAs and a library made from them. The DNA in these colonies can then be isolated for analysis.
If a plasmid vector is used, about 700,000 separate fragments are required to make a library of the human genome. By using bacteriophage λ, which can carry about four times as much DNA as a typical plasmid, the number of “volumes” in the library can be reduced to about 160,000. Although this still seems like a large number, a single petri plate can hold thousands of phage colonies, or plaques, which are easily screened for the presence of a particular DNA sequence by *hybridization to an appropriate nucleic acid probe.
*connect the concepts The technique of nucleic acid hybridization, illustrated in Figure 14.6, involves using a single-