DNA microarrays reveal RNA expression patterns
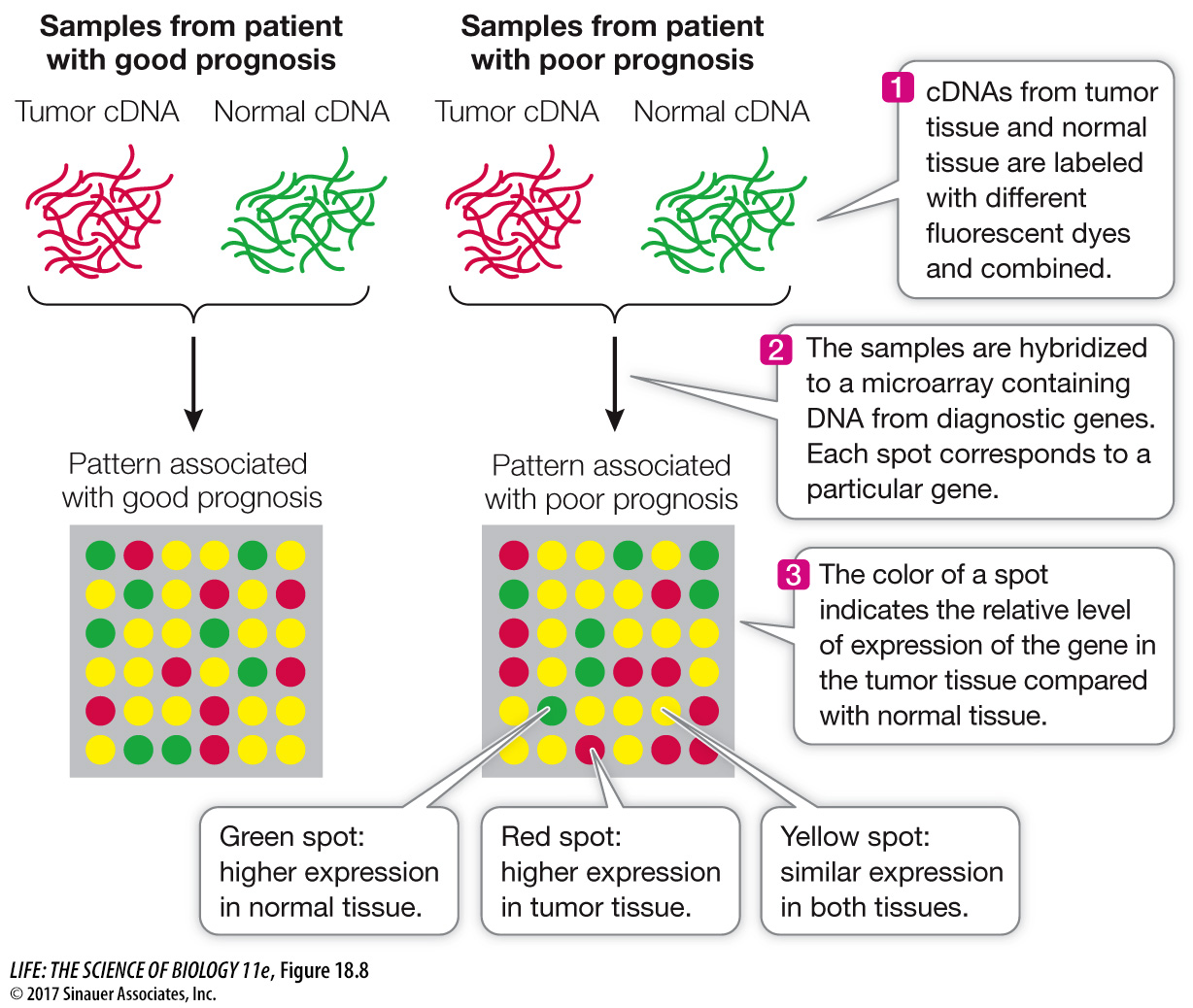
The science of genomics faces two major quantitative challenges. First, there are very large numbers of genes in eukaryotic genomes. Second, there are myriad distinct patterns of gene expression in different tissues at different times. For example, the cells of a skin cancer at its early stage may have a unique mRNA “fingerprint” that differs from those of normal skin cells and cells from a more advanced skin cancer. In such a case, the pattern of gene expression could provide invaluable information to a clinician trying to characterize a patient’s tumor (see below).
To identify distinct patterns of gene expression, scientists could isolate mRNA from cells and measure the amount of each gene’s mRNA one gene at a time by hybridization or RT-
DNA microarrays (“gene chips”) contain a series of DNA sequences attached to a glass slide in a precise order. The slide is divided into a grid of microscopic spots, or “wells.” Each spot contains thousands of copies of a particular oligonucleotide of 20 or more bases. A computer controls the addition of these oligonucleotide sequences in a predetermined pattern. Each oligonucleotide can hybridize with only one DNA or RNA sequence and thus is a unique identifier of a gene. Many thousands of different oligonucleotides can be placed on a single microarray to detect genetic variation. For example, DNA with a mutant allele may not hybridize to a target wild-
391
Animation 18.1 DNA Microarray Technology
www.life11e.com/