Reporter genes help select or identify host cells containing recombinant DNA
Even when a population of host cells is exposed to an appropriate vector, only a small proportion of the cells actually take up the vector. Furthermore, the process of making recombinant DNA is far from perfect. During a ligation reaction, DNA molecules can combine in various ways, many of which do not produce the desired recombinant molecule (see Figures 18.1 and 18.2). Methods have been developed to improve the chance that a desired combination will occur. A simple approach is to cut the vector with two different restriction enzymes that have sites near each other. This produces a molecule with incompatible sticky ends, which is much less likely to simply recircularize during the ligation reaction. The desired insert molecule is cut with the same two enzymes, so that theoretically a functional circular plasmid can be produced only if the vector and insert ligate with one another. Even so, it is often the case that only a small proportion of the ligation products have the desired recombinant sequence.
How can we identify or select the host cells that contain the desired sequence? One way is to use selectable markers, such as those for antibiotic resistance used in early experiments with recombinant DNA (see Figure 18.1). Selectable markers are one type of reporter gene, which is any gene whose expression is easily observed. Here are several types of reporter genes:
385
As described earlier, an antibiotic resistance gene in a plasmid or other vector allows the detection of transformed host cells grown in the presence of the selected antibiotic. If the host cells are normally sensitive to the antibiotic, they will grow on medium containing the antibiotic only if they have been transformed by the vector. This approach is used in the selection of transgenic prokaryotic and eukaryotic cells, including those of plants and animals.
The β-galactosidase (lacZ) gene in the E. coli lac operon (see Figure 16.4) codes for an enzyme that can convert the artificial substrate X-
Gal into a bright blue product. Many plasmids contain the lacZ gene with a multiple cloning site (i.e., multiple unique restriction enzyme sites where target DNA sequences can be inserted) within its sequence. These plasmids also carry genes for antibiotic resistance. Bacterial colonies containing the plasmid are selected on a solid medium containing the antibiotic. X- Gal is also included in the medium. If a bacterial colony contains a recombinant plasmid that carries the foreign DNA inserted into the lacZ gene, it will not make β-galactosidase and the colony will be white. Clones that contain the original plasmid with no insert express the lacZ gene and make blue colonies (Figure 18.3). research tools
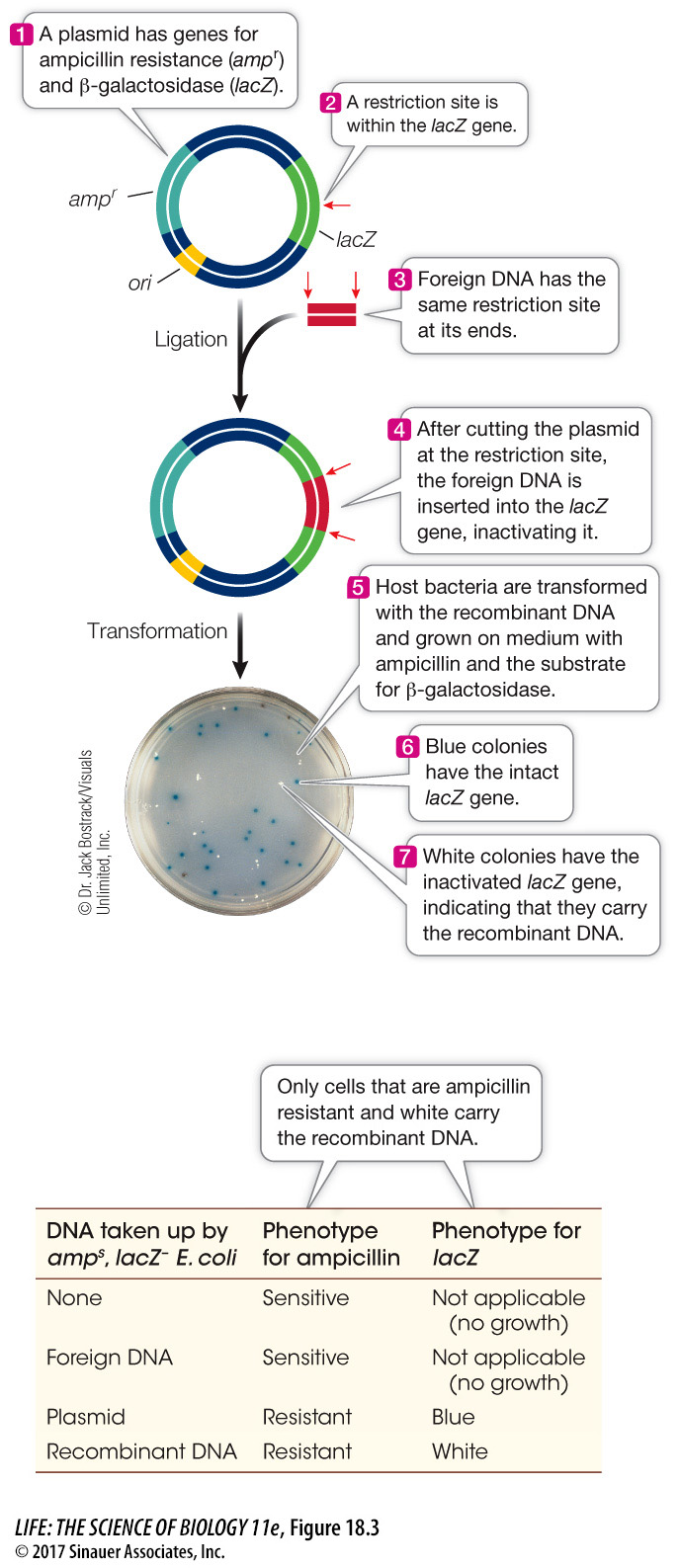 Figure 18.3 Selection for Recombinant DNA Selectable marker (reporter) genes are used by biologists to select for bacteria that have taken up a plasmid. In a typical experiment, most of the bacteria will not take up any DNA. Of those that do, only a small fraction will take up recombinant DNA.
Figure 18.3 Selection for Recombinant DNA Selectable marker (reporter) genes are used by biologists to select for bacteria that have taken up a plasmid. In a typical experiment, most of the bacteria will not take up any DNA. Of those that do, only a small fraction will take up recombinant DNA.Green fluorescent protein (GFP), which normally occurs in the jellyfish Aequorea victoria, emits visible green light when exposed to ultraviolet light. The gene for this protein has been isolated and incorporated into vectors. GFP is now widely used as a reporter gene (Figure 18.4). GFP has also been modified to emit other colors when exposed to ultraviolet light, and these new variants are widely used by molecular biologists.
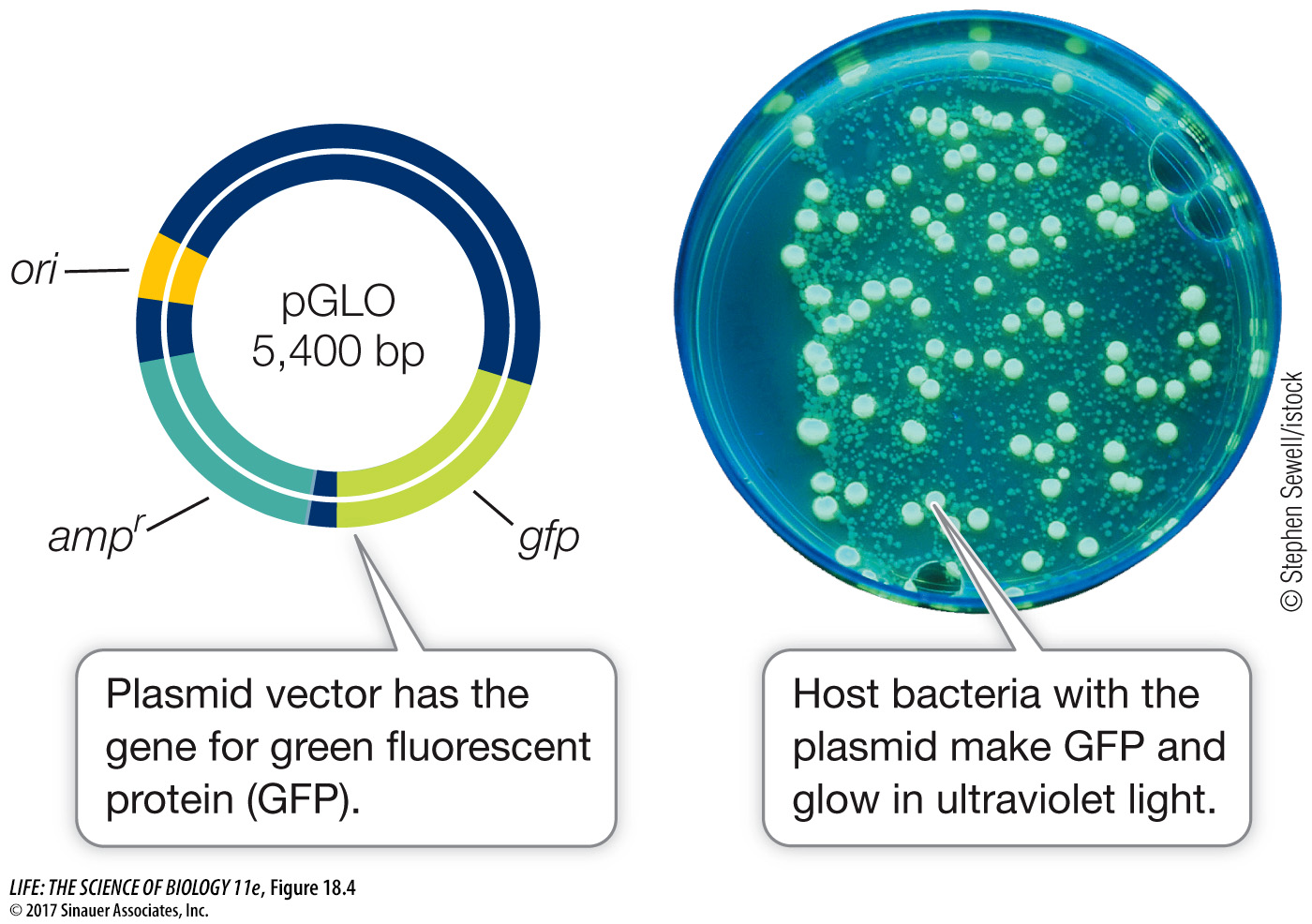 Figure 18.4 Green Fluorescent Protein as a Reporter The presence of a plasmid with the gene for green fluorescent protein (GFP) is readily apparent in transgenic cells because the GFP that the cells produce glows under ultraviolet light. This allows the identification of cells carrying a plasmid without the use of selection on antibiotics.
Figure 18.4 Green Fluorescent Protein as a Reporter The presence of a plasmid with the gene for green fluorescent protein (GFP) is readily apparent in transgenic cells because the GFP that the cells produce glows under ultraviolet light. This allows the identification of cells carrying a plasmid without the use of selection on antibiotics.Question
Q: Are cells killed during GFP selection? Why is this important?
Cells with GFP reporter plasmids are not killed. This means that the cell or organism with the plasmid can be grown directly, without killing other cells. Also, it means that live organism experiments, following the fates of cells with recombinant DNA over time, are possible.