Expression of transcription factor genes determines organ differentiation in plants
411
Like animals, plants have organs—
Activity 19.3 Genes and Development Simulation
www.life11e.com.ac19.3
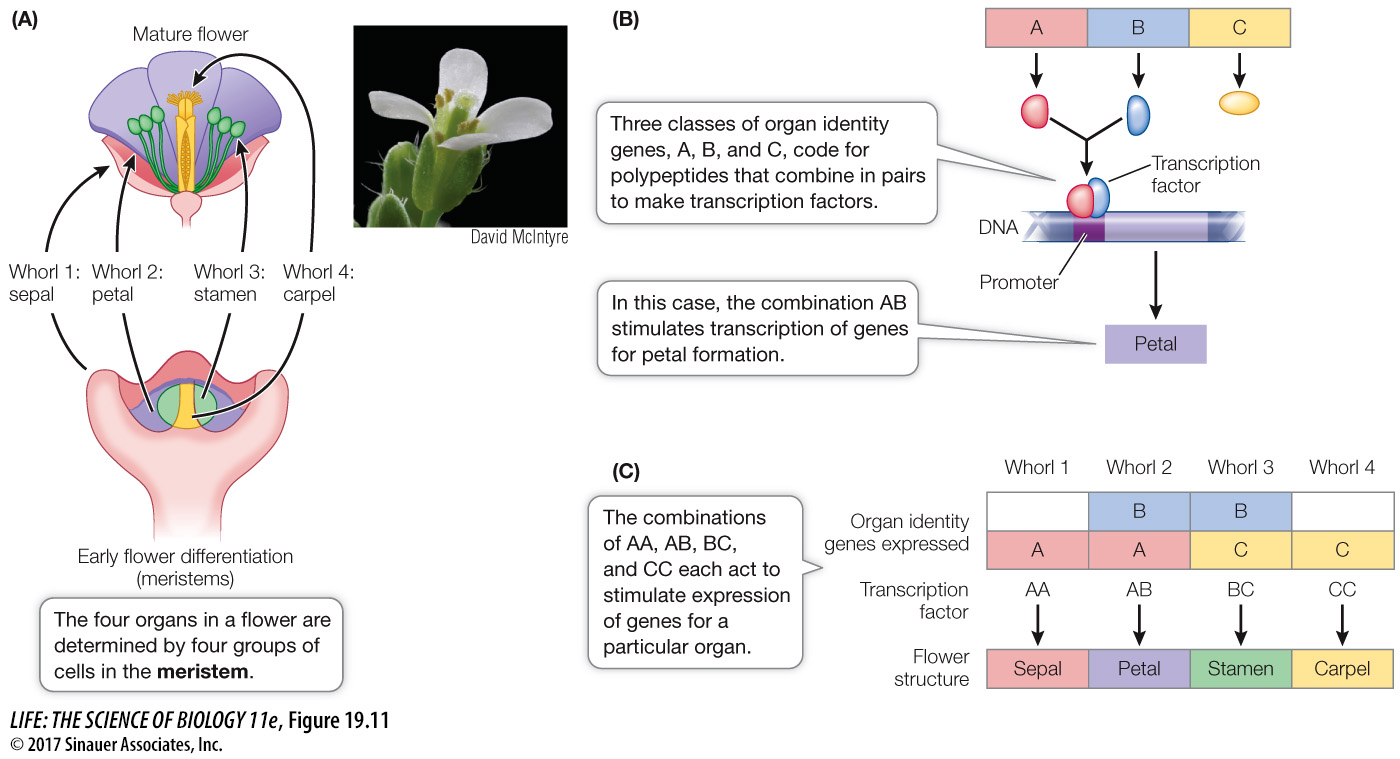
In Arabidopsis thaliana (thale cress), flowers develop in a radial pattern around the shoot (stem and leaf) apex as it develops and elongates. At the shoot apex and in other parts of the plant where growth and differentiation occur (such as the root tip), there are groups of undifferentiated, rapidly dividing cells called meristems. Each flower begins as a floral meristem of about 700 undifferentiated cells arranged in a dome, and the four whorls develop from this meristem. How is the identity of a particular whorl determined? Three classes of genes called organ identity genes encode proteins that act in combination to produce specific whorl features (Figure 19.11B and C):
Genes in class A are expressed in whorls 1 and 2 (which form sepals and petals, respectively).
Genes in class B are expressed in whorls 2 and 3 (which form petals and stamens).
Genes in class C are expressed in whorls 3 and 4 (which form stamens and carpels).
These genes encode transcription factors that are active as dimers, that is, proteins with two polypeptide subunits. The composition of the dimer determines which genes the transcription factor activates. For example, a dimer made up of two class A monomers activates transcription of the genes that make sepals; a dimer made up of A and B monomers results in petals, and so forth. A common feature of the A, B, and C proteins, as well as many other plant transcription factors, is a DNA-
Two lines of experimental evidence support this model for floral organ determination:
Loss-
of- function mutations: for example, a mutation in a class A gene results in no sepals or petals. 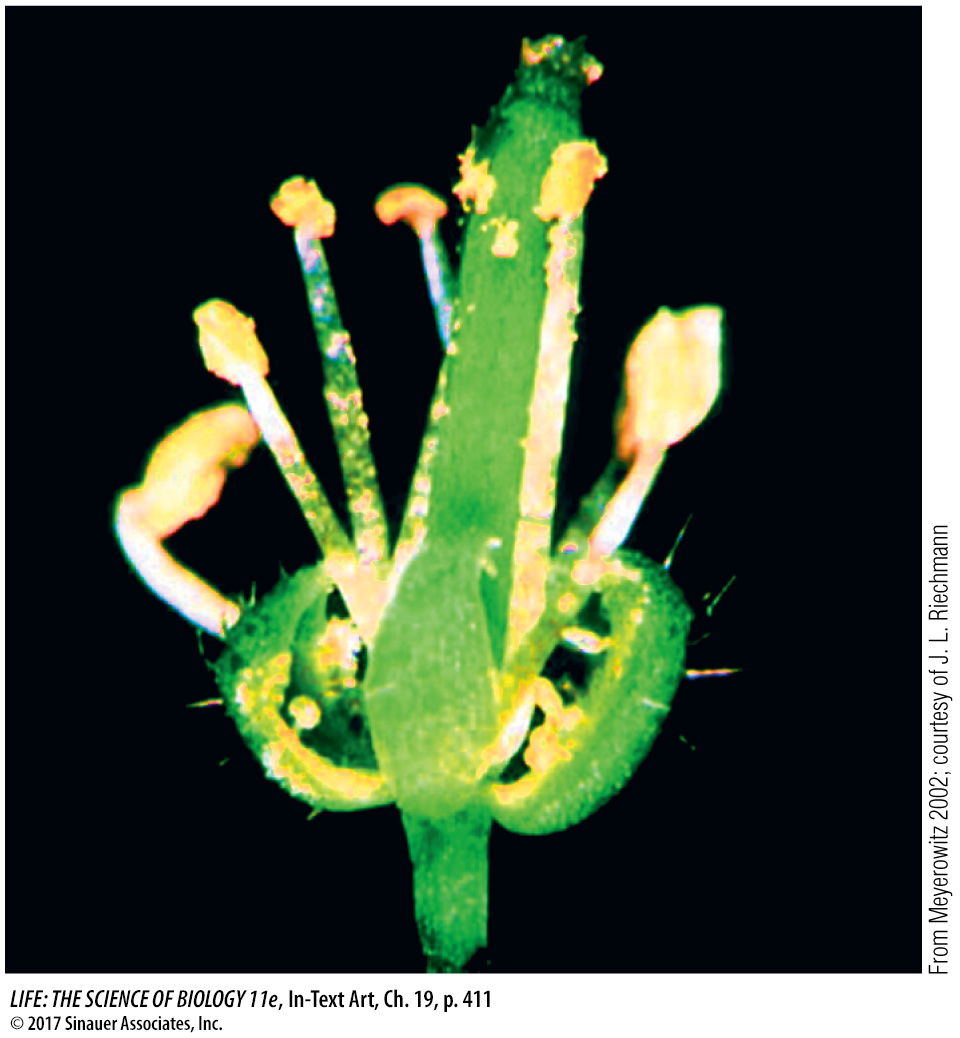
Gain-
of- function mutations: for example, a promoter for a class C gene can be artificially coupled to a class A gene. In this case, the class A gene is expressed in all four whorls, resulting in only sepals and petals. In any organism, the replacement of one organ for another is called homeosis, and this type of mutation is a homeotic mutation.
412

Transcription of the floral organ identity genes is controlled by other gene products, including the LEAFY protein. The wild-