Genome size also evolves
We know that genome size varies tremendously among organisms. Across broad taxonomic categories, there is some correlation between genome size and organismal complexity. The genome of the tiny bacterium Mycoplasma genitalium has only 470 genes. Rickettsia prowazekii, the bacterium that causes typhus, has 634 genes. Homo sapiens, by contrast, has about 21,000 protein-
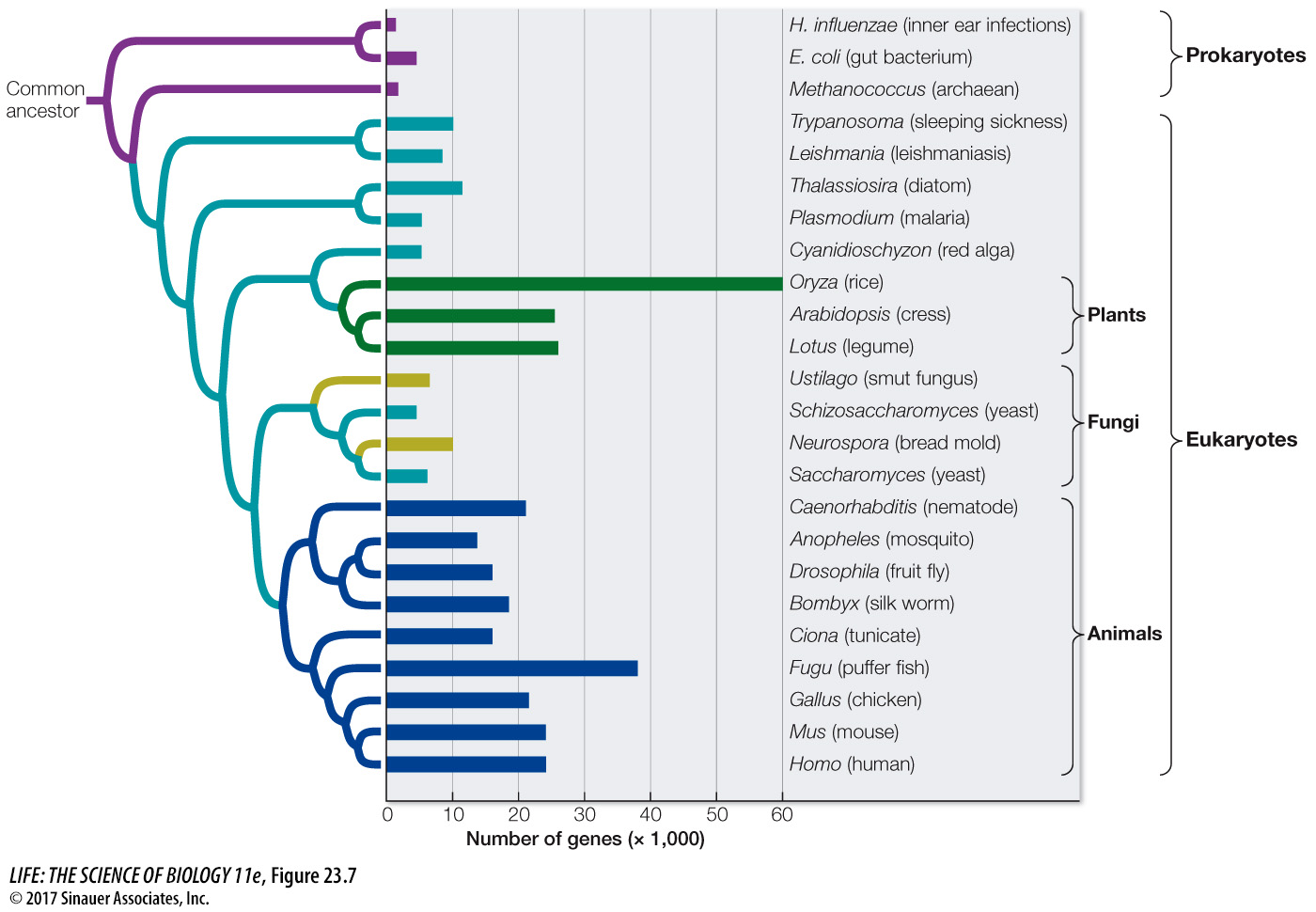
495
Differences in genome size are not so great if we take into account only the portion of the DNA that actually encodes RNAs or proteins. Although the organisms with the largest total amounts of nuclear DNA (some ferns and flowering plants) have 80,000 times as much DNA as do the bacteria with the smallest genomes, no species has more than about 100 times as many protein-
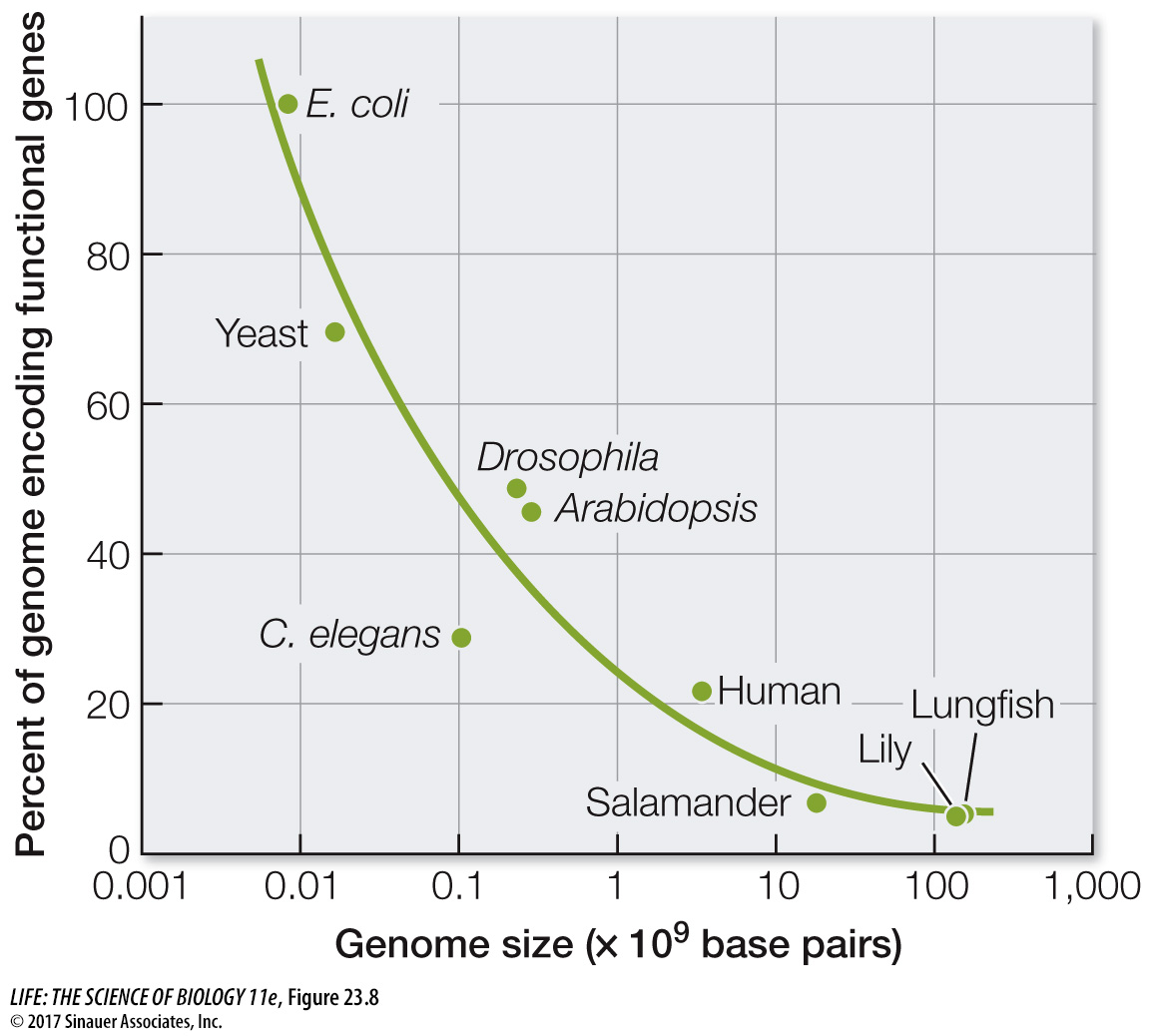
Why do the cells of most eukaryotic organisms have so much noncoding DNA? As we noted earlier, some of the noncoding DNA has a regulatory function that controls the degree or timing of expression coding genes. But the genomes of many species have far more noncoding DNA that is used for gene regulation. Does this extra noncoding DNA have a function, or is it “junk”? Many regions of noncoding DNA consist of pseudogenes (nonfunctional copies of former genes) that are carried in the genome simply because the cost of doing so is very small. These pseudogenes may become the raw material for the evolution of new genes with novel functions. Some noncoding DNA functions solely in maintaining chromosomal structure. Still other sequences consist of “selfish” transposable elements that proliferate because they reproduce faster than the host genome.
DNA does not just accumulate in genomes over time; noncritical nucleotide sequences are also lost from genomes. Some species differ so much in genome size because they lose noncritical sequences at very different rates. Investigators can use retrotransposons to estimate the rates at which species lose DNA. Retrotransposons are transposable elements (see Figure 17.4) that copy themselves through an RNA intermediate. The most common type of retrotransposon carries duplicated sequences at each end, called long terminal repeats, or LTRs. Occasionally, LTRs recombine in the host genome in such a way that the DNA between them is excised. When this happens, one recombined LTR is left behind. The number of such “orphaned” LTRs in a genome is a measure of how many retrotransposons have been lost. By comparing the number of LTRs in the genomes of Hawaiian crickets (Laupala) and fruit flies (Drosophila), investigators found that Laupala loses DNA more than 40 times more slowly than does Drosophila. Therefore it is not surprising that the genome of Laupala is much larger than that of Drosophila.
496
Why do species differ so greatly in the rate at which they gain or lose apparently functionless DNA? One hypothesis is that genome size is related to the rate at which the organism develops, which may be under selection pressure. Large genomes can slow down the rate of development and thus alter the relative timing of expression of particular genes. As discussed in Key Concept 19.4, changes in the timing of gene expression—
Another hypothesis is that the proportion of noncoding DNA is related primarily to population size. Noncoding sequences that are only slightly deleterious to the organism are likely to be purged by selection most efficiently in species with large population sizes. In species with small populations, the effects of genetic drift can overwhelm selection against noncoding sequences that have small deleterious consequences. Therefore selection against the accumulation of noncoding sequences is most effective in species with large populations, and such species (such as bacteria and yeasts) have relatively little noncoding DNA compared with species with small populations (see Figure 23.9).