recap
23.2 recap
The neutral theory of molecular evolution provides an explanation for the relatively constant rate of molecular change seen in many species. By examining the relative rates of synonymous and nonsynonymous substitutions in genes over time, biologists can distinguish the evolutionary mechanisms acting on individual genes. The noncoding portion of eukaryote genomes is more variable in size across species than is the coding portion.
learning outcomes
You should be able to:
Explain why substitutions may be neutral or selective in a given organism.
Provide a mathematical explanation for why the rate of fixation of neutral mutations is independent of population size.
Evaluate gene sequences of protein-
coding genes to identify codons that are evolving under purifying versus positive selection, and apply this knowledge to a biological problem. Describe the evolution of genomes in a way that incorporates consideration of noncoding DNA.
Question 1
Over evolutionary history, many groups of organisms that inhabit caves have lost the organs of sight. For example, although surface-
This problem can be investigated by sequencing and comparing the genes for opsins in surface-
Question 2
Why is the rate of fixation of neutral mutations independent of population size?
A given neutral mutation will arise more often in a large population than in a small population, but any mutation that does arise is more likely to be fixed in a small population than in a large population. These two influences of population size exactly cancel each other out, so that overall, the rate of fixation of neutral mutations depends only on the mutation rate and is independent of population size.
In a diploid population of size N and a neutral mutation rate μ per gamete per generation at a locus, the number of new mutations would be, on average, 2Nμ, because 2N gene copies are available to mutate. The probability that a given mutation will be fixed by drift alone is its frequency, which equals 1/(2N) for a newly arisen mutation. We can multiply these two terms to get the rate of fixation of neutral mutations (m) in a given population of N diploid individuals: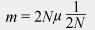
But we can simplify this equation by canceling out 2N on the right side of the equation: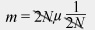
This just leaves m = μ. In words, the rate of fixation of neutral alleles is equal to the mutation rate of neutral alleles and independent of population size.
Question 3
An investigator compared many gene sequences encoding surface proteins from influenza viruses sampled over time and collected the data shown in the table below.
| Codon position | Synonymous substitutions | Nonsynonymous substitutions |
|---|---|---|
| 12 | 0 | 7 |
| 15 | 1 | 9 |
| 61 | 0 | 12 |
| 80 | 7 | 0 |
| 137 | 12 | 1 |
| 156 | 24 | 2 |
| 165 | 3 | 4 |
| 226 | 38 | 3 |
Which positions encode amino acids that are likely to have changed as a result of positive selection? Explain your answer.
497
Which positions encode amino acids that are likely to have changed as a result of purifying selection? Explain your answer.
(Hint: To calculate rates of each substitution type, consider the number of synonymous and nonsynonymous substitutions present relative to the number of possible substitutions of each type. There are approximately three times as many possible nonsynonymous substitutions as there are synonymous substitutions.)
(a) Codon numbers 12, 15, and 61 are likely to be evolving under positive selection for change because each of these codons has experienced a higher rate of nonsynonymous substitutions (which give rise to amino acid replacements) than synonymous substitutions.
(b) Codon numbers 80, 137, 156, and 226 are likely evolving under purifying selection, as the vast majority of changes at these codons are synonymous substitutions, which do not result in amino acid replacements. Substitutions that result in amino acid changes (nonsynonymous substitutions) undoubtedly occur but are usually selected against in the population. Codon number 165 has experienced similar numbers of synonymous and nonsynonymous substitutions. There are approximately three times as many possible nonsynonymous substitutions as there are synonymous substitutions. Therefore the number of synonymous substitutions is slightly higher than expected if the rates of each type of substitution are equal. Codon 165 may be evolving under weak purifying selection; it is the codon that is closest to neutral among the codons shown in the table.
Question 4
Suggest and contrast two hypotheses for the wide diversity of genome sizes among different organisms.
Example hypothesis 1: Genome size varies primarily because noncoding DNA can effect changes in gene expression, and many species have evolved largely through changes in gene expression.
Example hypothesis 2: Genome size varies primarily as a function of population size. Species with small population sizes may accumulate large amounts of noncoding DNA because of weak selection against accumulation of slightly deleterious “junk” DNA in small populations.
Contrasting the two hypotheses: If hypothesis 1 explains most of the variation in genome size among organisms, then we would expect genome size to be largely independent of population size, which is not the case. For example, Figure 23.9 indicates that the species with the largest genomes, like the lungfish, generally have much smaller population sizes than the species with the smallest genomes, like the E. coli or yeast. However, both hypotheses may explain some of the variation in genome size, and careful, controlled experiments would be needed to determine how much of the variation in genome size is explained by the effects of selection (hypothesis 1) versus population size (hypothesis 2).
We have examined some of the ways in which organisms can lose DNA without losing gene functions. But how do organisms gain new functions through time?