Molecular sequence data are used to determine the evolutionary history of genes
A gene tree can show the evolutionary relationships of a single gene in different species, or it can trace the evolution of members of a gene family (as in Figure 23.11). The methods for constructing a gene tree are the same as those we described in Key Concept 21.2 for building phylogenetic trees of species. The process involves identifying differences between genes and using those differences to reconstruct the evolutionary history of the genes. Gene trees are often used to construct phylogenetic trees of species, but the two types of trees are not necessarily equivalent. Processes such as gene duplication can give rise to differences between the phylogenetic trees of genes and species. From a gene tree, biologists can reconstruct the history and timing of gene duplication events and learn how gene diversification has resulted in the evolution of new protein functions.
501
All the genes of a particular gene family have similar sequences because they have a common ancestry. As we discussed in Key Concept 21.1, features that are similar as a result of common ancestry are said to be homologous. When discussing gene trees, however, we usually need to distinguish between two forms of homology. Homologous genes that are found in different species and whose divergence we can trace to the speciation events that gave rise to those species are called orthologs. Homologous genes in the same or different species that are related through gene duplication events are called paralogs. When we examine a gene tree, the questions we wish to address determine whether we should compare orthologous or paralogous genes. If we wish to reconstruct the evolutionary history of the species that contain the genes, then our comparison should be restricted to orthologs (because they will reflect the history of speciation events). If we are interested in the changes in function that have resulted from gene duplication events, however, then the appropriate comparison is among paralogs (because they will reflect the history of gene duplication events). If our focus is on the diversification of a gene family through both processes, then we will want to include both paralogs and orthologs in our analysis.
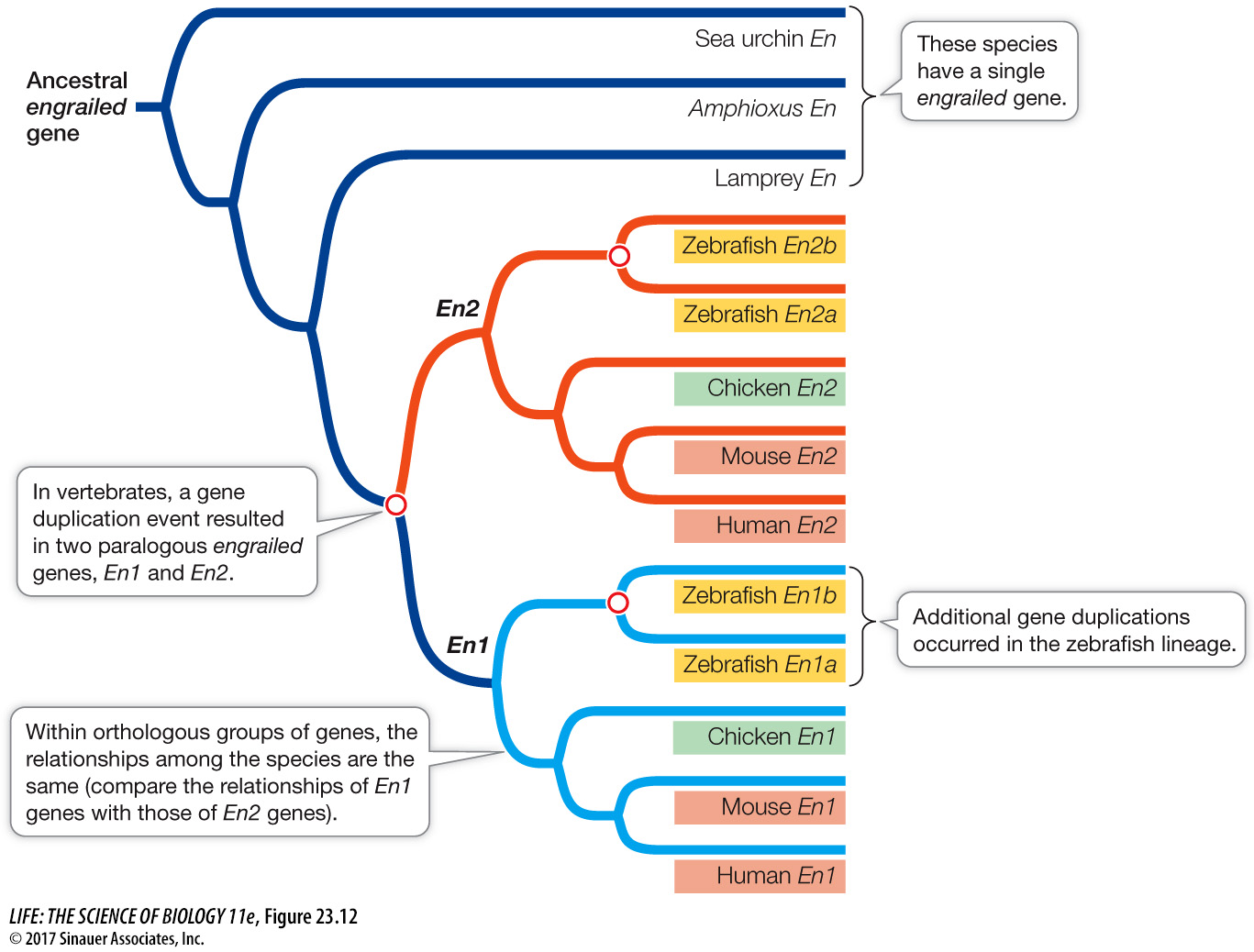
Figure 23.12 depicts a gene tree for the members of a gene family called engrailed (its members encode transcription factors that regulate development). At least three gene duplications have occurred in this family, resulting in up to four different engrailed genes (En) in some vertebrate species (such as the zebrafish). All of the engrailed genes are homologs because they have a common ancestor. Gene duplication events have generated paralogous engrailed genes (En1 and En2) in some lineages of vertebrates. We could compare the orthologous sequences of the En1 group of genes to reconstruct the history of the bony vertebrates (i.e., all the vertebrate species in Figure 23.12 except the lamprey), or we could use the orthologous sequences of the En2 group of genes and expect the same answer (because there is only one history of the underlying speciation events). All bony vertebrates have both groups of engrailed genes because the two groups arose from a gene duplication event in the common ancestor of bony vertebrates. If we wanted to focus on the diversification that occurred as a result of this duplication, then the appropriate comparison would be between the paralogous genes of the En1 versus En2 groups.
Question
Q: How many gene duplication events occurred just within the zebrafish lineage?
Two gene duplication events were restricted to the zebrafish lineage: the event that gave rise to the zebrafish En1a and En1b genes, and the event that gave rise to the zebrafish En2a and En2b genes.