In vitro evolution is used to produce new molecules
As biologists studied the relationships among selection, evolution, and function in macromolecules, they realized that molecular evolution could be used in a controlled laboratory environment to produce new molecules with novel and useful functions. Thus were born the applications of in vitro evolution.
Living organisms produce thousands of compounds that humans have found useful. The search for such naturally occurring compounds, which can be used for pharmaceutical, agricultural, or industrial purposes, has been termed “bioprospecting.” These compounds are the result of millions of years of molecular evolution across millions of species of living organisms. Yet biologists can also imagine molecules that could have evolved but, lacking the right combination of selection pressures and opportunities, have not. For instance, we might want to have a molecule that binds a particular environmental contaminant so that the contaminant can be isolated and extracted from the environment. But if the contaminant is synthetic (i.e., not produced naturally), it is unlikely that any living organism will have evolved a molecule with the function we desire. This problem was the inspiration for the field of in vitro evolution.
The principles of in vitro evolution are based on the principles of molecular evolution that we have learned from the natural world. Consider the evolution of a new RNA molecule that was produced in the laboratory using the principles of mutation and selection. This molecule’s intended function was to join two other RNA molecules. That is, the lab created a ribozyme—
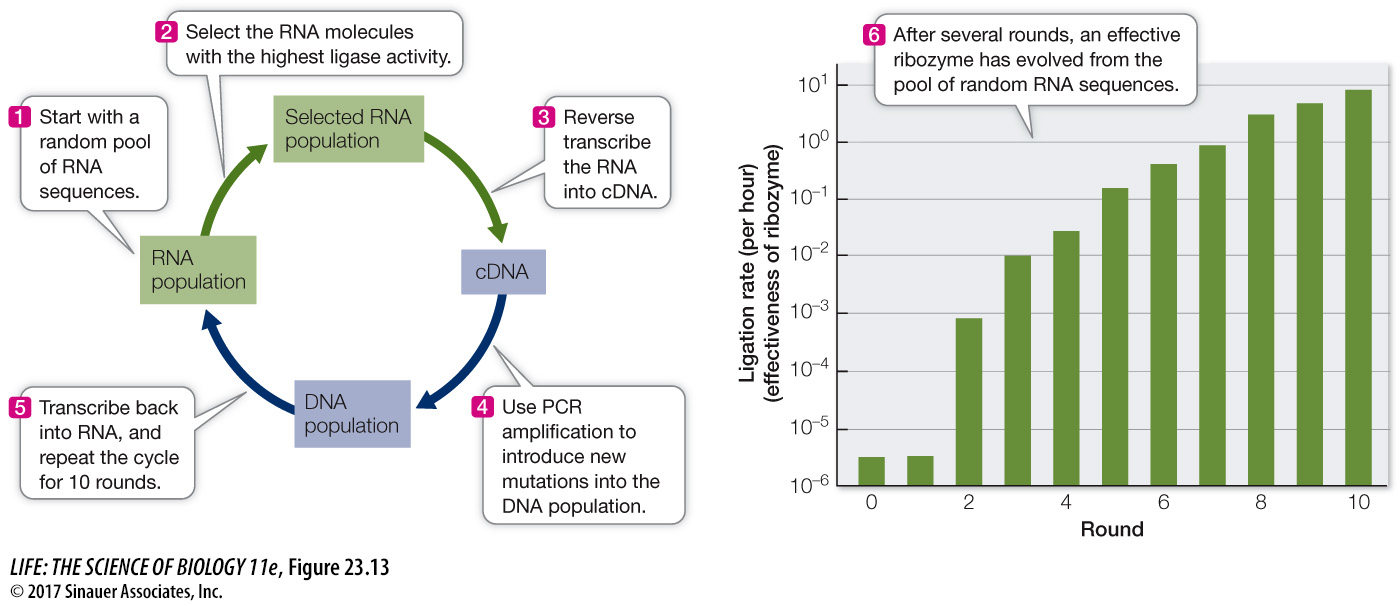
PCR amplification is not perfect, and it introduced many new mutations into the pool of sequences. These sequences were then transcribed back into RNA molecules using RNA polymerase, and the process was repeated. The ligase activity of the RNAs evolved quickly; after 10 rounds of in vitro evolution, it had increased by about 7 million times. Similar techniques have since been used to create a wide variety of molecules with novel enzymatic and binding functions.