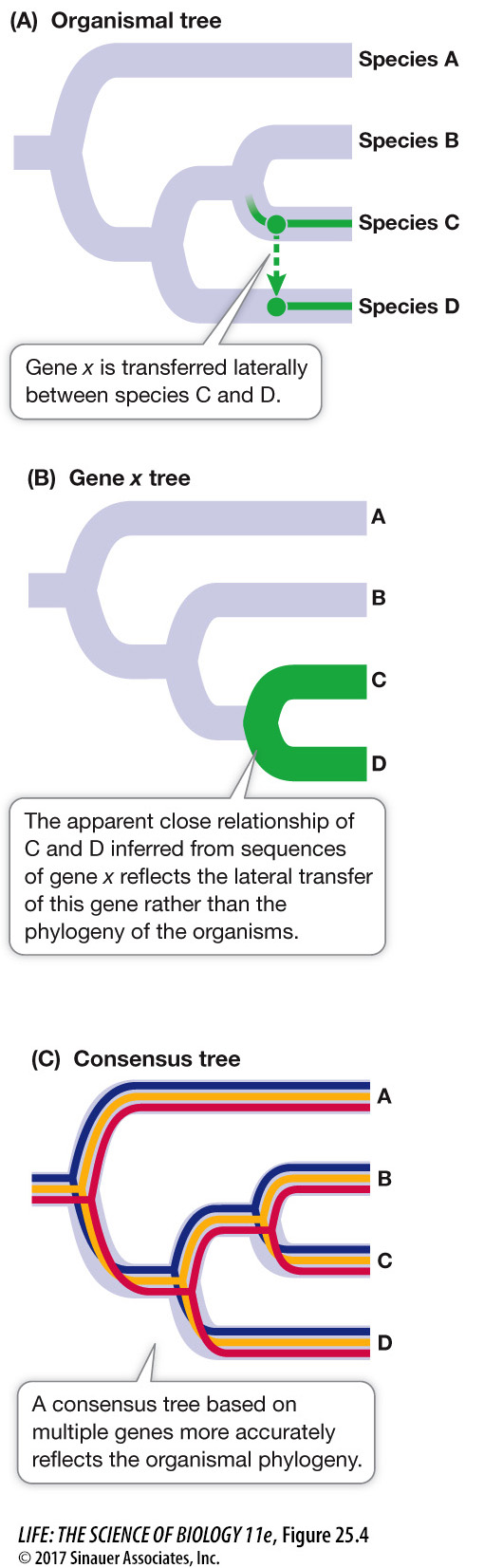
Lateral gene transfer can lead to discordant gene trees
As noted earlier, prokaryotes reproduce by binary fission. If we could follow these divisions back through evolutionary time, we would be tracing the complete tree of life. At a much broader scale, these divisions of organisms lead to splits among the major evolutionary lineages, or species of life (represented in highly abbreviated form in Appendix A). Because binary fission is an asexual process that replicates whole genomes, we would expect phylogenetic trees of prokaryotes constructed from most gene sequences (see Chapter 22) to reflect these same relationships.
Even though binary fission is an asexual process, there are other processes—
*connect the concepts Prokaryote exchange of genetic material by conjugation and transformation is described in Key Concepts 12.6 and 13.1, respectively.
From early in evolution to the present day, some genes have been moving “sideways” from one prokaryote species to another, a phenomenon known as lateral gene transfer (sometimes called horizontal gene transfer). Lateral gene transfers are well documented among closely related species, and some have been documented even across the domains of life.
Consider, for example, the genome of Thermotoga maritima, a bacterium that can survive extremely high temperatures. By comparing the 1,869 gene sequences of T. maritima with sequences encoding the same proteins in other species, investigators found that some of this bacterium’s genes have their closest relationships not with the genes of other bacterial species, but with the genes of archaea that live in similar extreme environments.
When genes involved in lateral transfer events are sequenced and analyzed, the resulting gene trees will not match the organismal tree in every respect (Focus: Key Figure 25.4). The individual gene trees will vary because the history of lateral transfer events is different for each gene. Biologists can reconstruct the underlying organismal phylogeny by comparing multiple genes (to produce a consensus tree) or by concentrating on genes that are unlikely to be involved in lateral gene transfer events. For example, genes that are involved in fundamental cellular processes (such as the rRNA genes discussed above) are unlikely to be replaced by the same genes from other species because functional, locally adapted copies of these genes are already present.
focus: key figure
Question
Q: Why are multiple lateral gene transfers between the same two branches on a phylogeny expected to be rare, at least compared with similarities inherited through the stable core?
Lateral gene transfers are most likely to occur when there is a selective advantage to the transfer. One species may receive genes from many others, but the chances are smaller that a substantial portion of the genome of one species will be transferred into another. In contrast, the stable core of genes controls critical metabolic functions of the cell, so all of these genes are expected to be inherited from ancestors to descendants together.
What kinds of genes are most likely to be involved in lateral gene transfer? Genes that result in a new adaptation that confers higher fitness on a recipient species are most likely to be transferred repeatedly among species. For example, genes that produce antibiotic resistance are often transferred among bacterial species on plasmids, especially under the strong selection pressure such as that imposed by modern antibiotic medications. Improper or overly frequent use of antibiotics can select for resistant strains of bacteria that are much harder to treat. This selection for antibiotic resistance explains why informed physicians have become more careful in prescribing antibiotics.
It is debatable whether lateral gene transfer has seriously complicated our attempts to resolve the tree of prokaryotic life. Recent work suggests that it has not. Lateral gene transfer rarely creates problems at higher taxonomic levels, even though it may complicate our understanding of the relationships among individual species. Some species clearly obtain some of their genes from otherwise distantly related species, so evolutionary histories of individual genes may differ within a single organism. But it is now possible to make nucleotide sequence comparisons involving entire genomes, and these studies are revealing a stable core of crucial genes that are uncomplicated by lateral gene transfer. Gene trees based on this stable core more accurately reveal the organismal phylogeny (see Figure 25.4). The problem remains, however, that only a very small proportion of the prokaryotic world has been described and studied.