RNA Processing Solves the Problem of Pervasive Transcription of the Genome in Metazoans
As discussed in Chapter 9, analysis of the location of transcribing RNA polymerase II in metazoan cells revealed the surprising result that the polymerase transcribes in the downstream direction, into coding regions, and in the upstream direction, away from coding regions, at nearly equal frequency from most promoters (see Figure 9-18). This finding was confirmed by deep sequencing of small RNAs isolated from metazoan cells, which revealed low levels of short, capped RNAs transcribed from both the sense and antisense strands at CpG island promoters, which account for some 70 percent of mammalian promoters. Indeed, deep sequencing of all cellular RNAs showed that both strands of nearly the entire genome are transcribed, although much of the resulting RNA is present at extremely low concentrations of less than one molecule per cell. This finding raised the question of how the cell deals with such “pervasive transcription.”
Sequence analysis of these low-abundance short, capped RNAs indicates that they are probably prevented from reaching high concentrations by RNA processing and nuclear surveillance for abnormally processed RNAs. Sequencing of RNAs from several cell types has revealed that the antisense RNAs have a higher frequency of AAUAAA polyadenylation signal sequences transcribed from the AT-rich DNA of most metazoans (~60 percent AT in mammals) than do transcripts transcribed in the sense direction into coding regions. Because of the high AT composition of mammalian DNA, an AAUAAA sequence in an antisense transcript is frequently followed by a U-rich sequence that may function as the downstream element of a bona fide pre-mRNA cleavage/polyadenylation signal (see Figure 10-15). These cleavage/polyadenylation signals occur much less frequently in transcripts going into coding regions. Where they do occur in the sequence of pre-mRNAs, in either exons or introns, they usually lie downstream of consensus base-pairing sites for U1 snRNA, which has been found to suppress cleavage/polyadenylation following nearby AAUAAA sequences. This function of U1 snRNA may help to explain why the U1 snRNP is much more abundant than the other spliceosomal snRNPs. This is not the case for cleavage/polyadenylation signals used in the processing of 3′ ends of mRNAs because U1 snRNA associates with the 5′ end of the terminal intron, far from the poly(A) site. In addition, as discussed above, transcription by RNA polymerase II usually terminates within ~2 kb following cleavage and polyadenylation of a pre-mRNA. Consequently, the enrichment of poly(A) sites, and the relative lack of binding sites for U1 snRNA, in antisense transcripts may lead to cleavage of most of these transcripts within ~2 kb of the transcription start site by cleavage/polyadenylation factors (see Figure 10-15), followed by termination of transcription (Figure 10-17). Cleaved antisense transcripts are probably degraded by the same nuclear exonucleases that degrade introns spliced out of pre-mRNAs and sequences downstream of pre-mRNA cleavage/polyadenylation sites, as well as sequences processed out of rRNA and tRNA precursors, discussed in a later section (see Figure 10-1). As a result, even though a large number of polymerases transcribe in the “wrong” direction, most of the transcripts generated in this way are rapidly degraded.
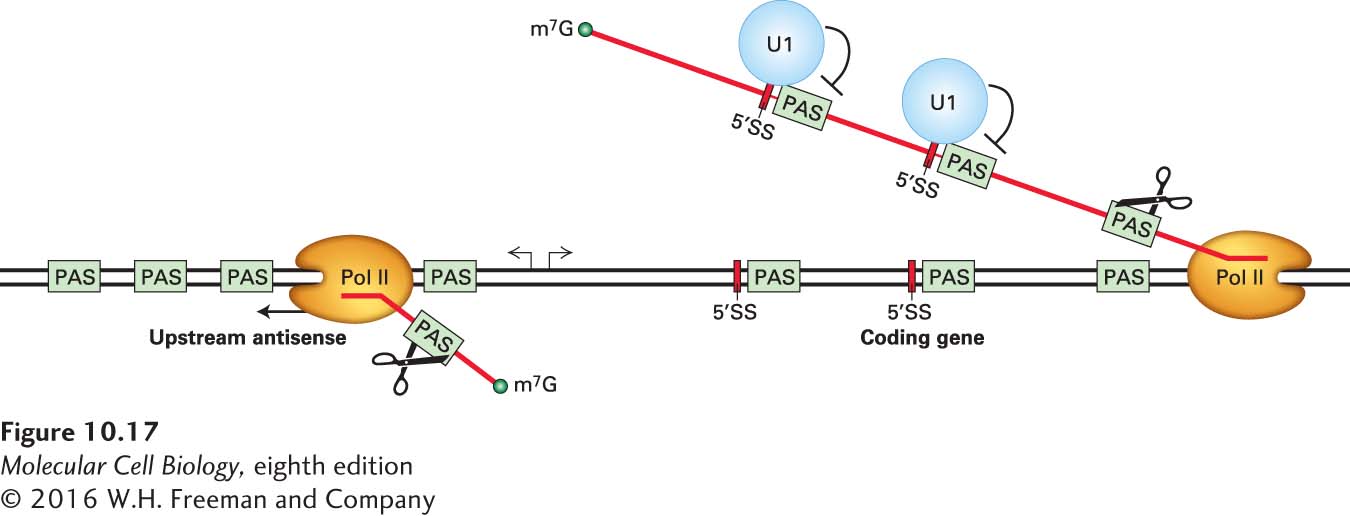
FIGURE 10-17 RNA transcribed in the "wrong" direction from most promoters in metazoans has a high frequency of polyadenylation signals and a low frequency of binding sites for U1 snRNA. This pattern may account for the termination of transcription in the "wrong" direction after about 2 kb for most of these transcripts. PAS represents polyadenylation signals encoded in the DNA that are transcribed into RNA. Cleavage of transcripts transcribed in the upstream direction (scissors) is proposed to generate free RNA ends that are digested by the nuclear exosome and a nuclear 5′→3′ exonuclease, XRN1. In contrast, pre-mRNAs synthesized by RNA polymerase II transcribing into coding regions have evolved to have few polyadenylation signals. Where they do occur, these signals are usually preceded by a binding site for U1 snRNP, which inhibits cleavage at a nearby PAS (stop sign). However, the PAS used to generate the 3′ end of an mRNA does not have a closely associated U1 RNP binding site. See A. E. Almada et al., 2013, Nature 499:360.
