RNA Editing Alters the Sequences of Some Pre-mRNAs
In the mid-1980s, sequencing of numerous cDNA clones and corresponding genomic DNAs from multiple organisms led to the unexpected discovery of another type of pre-mRNA processing. In this type of processing, called RNA editing, the sequence of a pre-mRNA is altered; as a result, the sequence of a mature mRNA differs from that of the exons encoding it in genomic DNA.
RNA editing is widespread in the mitochondria of protozoans and plants as well as in chloroplasts. In the mitochondria of certain pathogenic trypanosomes, more than half the sequence of some mRNAs is altered from the sequence of the corresponding primary transcripts. Additions and deletions of specific numbers of Us follow templates provided by base-paired short “guide” RNAs. These RNAs are encoded by thousands of small circular DNA molecules concatenated to many fewer large DNA molecules. The reason for this baroque mechanism for encoding mitochondrial proteins in such protozoans is not clear. But this system does represent a potential target for drugs to inhibit the complex processing enzymes essential to the microbe that do not exist in the cells of its human or other vertebrate hosts.
In higher eukaryotes, RNA editing is much rarer, and thus far, only single-base changes have been observed. Such minor editing, however, turns out to have significant functional consequences in some cases. An important example of RNA editing in mammals involves the APOB gene, which encodes two alternative forms of a serum protein that is central to the uptake and transport of cholesterol. Consequently, it is important in the pathogenic processes that lead to atherosclerosis, the arterial disease that is the major cause of death in the developed world. The APOB gene encodes both the serum protein apolipoprotein B-100 (apoB-100), which is expressed in hepatocytes, the major cell type in the liver, and apoB-48, which is expressed in intestinal epithelial cells. The 240-kDa apoB-48 corresponds to the N-terminal region of the 500-kDa apoB-100. Both ApoB proteins are components of the large lipoprotein complexes we described in Chapter 7, which transport lipids in the serum. However, only low-density lipoprotein (LDL) complexes, which contain apoB-100 on their surface, deliver cholesterol to body tissues by binding to the LDL receptor that is present on all cells (see Figures 14-27 and 14-29).
The cell-type-specific expression of the two forms of ApoB results from editing of ApoB pre-mRNA so as to change the nucleotide at position 6666 in the sequence from a C to a U. This alteration, which occurs only in intestinal cells, converts a CAA codon for glutamine to a UAA stop codon, leading to synthesis of the shorter apoB-48 (Figure 10-22). Studies with the partially purified enzyme that performs the post-transcriptional deamination of C6666 to U (see Figure 2-17) shows that it can recognize and edit an RNA as short as 26 nucleotides containing the sequence surrounding C6666 in the ApoB primary transcript.
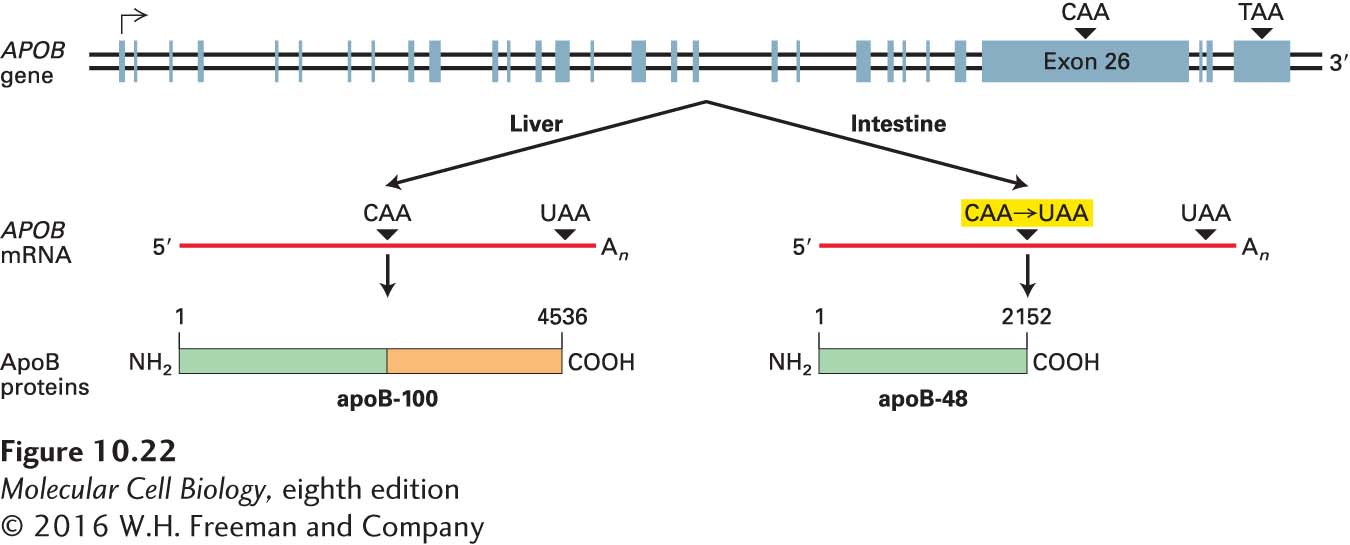
FIGURE 10-22 RNA editing of APOB pre-mRNA. The APOB mRNA produced in the liver has the same sequence as the exons in the primary transcript. This mRNA is translated into apoB-100, which has two functional domains: an N-terminal domain (green) that associates with lipids and a C-terminal domain (orange) that binds to LDL receptors on cell membranes. In the APOB mRNA produced in the intestine, however, the CAA codon in exon 26 is edited to a UAA stop codon. As a result, intestinal cells produce apoB-48, which corresponds to the N-terminal domain of apoB-100. See P. Hodges and J. Scott, 1992, Trends Biochem. Sci. 17:77.
