Sequence-Specific RNA-Binding Proteins Control Translation of Specific mRNAs
In contrast to the global mRNA regulation we have just described, other mechanisms have evolved for controlling the translation of certain specific mRNAs. These mechanisms usually rely on sequence-specific RNA-binding proteins that bind to a particular sequence or structure in the mRNA. When such proteins bind to the 5′ UTR of an mRNA, the small ribosomal subunit's ability to scan to the first initiation codon is blocked, inhibiting translation initiation. Binding in other regions can either promote or inhibit mRNA degradation.
Control of intracellular iron concentrations by the iron-response element–binding protein (IRE-BP) is an elegant example of a system in which a single protein regulates the translation of one mRNA and the degradation of another. Precise regulation of cellular iron ion concentrations is critical to the cell. Multiple enzymes and proteins contain Fe2+ as a cofactor, such as enzymes of the citric acid cycle (see Figure 12-16) and electron-carrying proteins involved in the generation of ATP by mitochondria and chloroplasts (see Chapter 12). On the other hand, excess Fe2+ generates free radicals that react with and damage cellular macromolecules. When intracellular iron stores are low, a dual-control system operates to increase the level of cellular iron; when iron is in excess, the system operates to prevent accumulation of toxic levels of free ions.
One component of this system is regulation of the production of ferritin, an intracellular protein that binds and stores excess cellular iron. The 5′ UTR of ferritin mRNA contains iron-response elements (IREs) that have a stem-loop structure. IRE-BP recognizes five specific bases in the IRE loop and the duplex nature of the stem. At low iron concentrations, IRE-BP is in an active conformation that binds to the IREs (Figure 10-33a). The bound IRE-BP blocks the small ribosomal subunit from scanning for the AUG start codon (see Figure 5-23), thereby inhibiting translation initiation. The resulting decrease in ferritin means that less iron is complexed with ferritin, and therefore more iron is available to iron-requiring enzymes. At high iron concentrations, IRE-BP is in an inactive conformation that does not bind to the 5′ IREs, so translation initiation can proceed. The newly synthesized ferritin then binds free iron ions, preventing their accumulation to harmful levels.
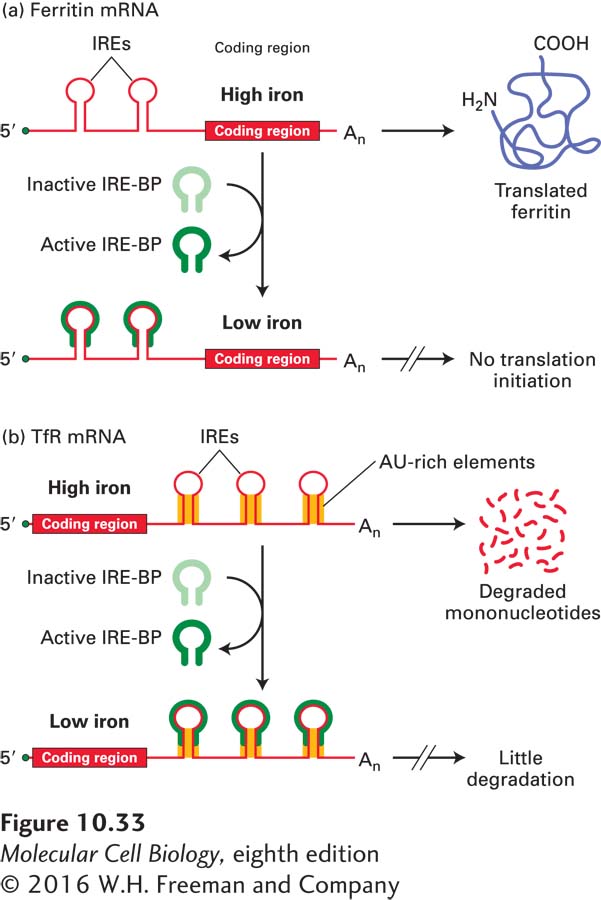
FIGURE 10-33 Iron-dependent regulation of mRNA translation and degradation. The iron-response element–binding protein (IRE-BP) controls (a) translation of ferritin mRNA and (b) degradation of transferrin-receptor (TfR) mRNA. At low intracellular iron concentrations, IRE-BP binds to iron-response elements (IREs) in the 5′ or 3′ UTR of these mRNAs. At high iron concentrations, IRE-BP undergoes a conformational change and cannot bind either mRNA. The dual control by IRE-BP precisely regulates the level of free iron ions within cells. See the text for discussion.
The other part of this regulatory system controls the import of iron into cells. In vertebrates, ingested iron is carried through the circulatory system bound to a protein called transferrin. After binding to the transferrin receptor (TfR) in the plasma membrane, the transferrin-iron complex is brought into cells by receptor-mediated endocytosis (see Figure 14-31). The 3′ UTR of TfR mRNA contains IREs whose stems have destabilizing AU-rich elements (Figure 10-33b). At high iron concentrations, when IRE-BP is in its inactive, nonbinding conformation, these AU-rich elements promote degradation of TfR mRNA by the mechanism described earlier in this section that leads to rapid degradation of other short-lived mRNAs with AU-rich elements. The resulting decrease in production of the transferrin receptor quickly reduces iron import, thus protecting the cell from excess iron. At low iron concentrations, however, IRE-BP is active and can bind to the 3′ IREs in TfR mRNA. The bound IRE-BP blocks recognition of the AU-rich elements by the proteins that would otherwise lead to rapid degradation of the mRNAs. As a result, production of the transferrin receptor increases, and more iron is transported into the cell.
Other regulated RNA-binding proteins function to control the translation or degradation of specific mRNAs in a similar manner. For example, a heme-sensitive RNA-binding protein controls translation of the mRNA encoding aminolevulinate (ALA) synthase, a key enzyme in the synthesis of heme. Similarly, in vitro studies have shown that the mRNA encoding the milk protein casein is stabilized by the hormone prolactin and rapidly degraded in its absence.
