Amphipathic N-Terminal Targeting Sequences Direct Proteins to the Mitochondrial Matrix
All proteins that travel from the cytosol to the same mitochondrial destination have targeting signals that share common motifs, although the signal sequences are not identical. Thus the receptors that recognize such signals are able to bind to a number of different but related sequences. The most extensively studied sequences for localizing proteins to mitochondria are the matrix-targeting sequences. These sequences, located at the N-terminus, are usually 20–50 amino acids in length. They are rich in hydrophobic amino acids, positively charged basic amino acids (arginine and lysine), and hydroxylated ones (serine and threonine), but tend to lack negatively charged acidic residues (aspartate and glutamate).
Mitochondrial matrix–targeting sequences are thought to assume an α-helical conformation in which positively charged amino acids predominate on one side of the helix and hydrophobic amino acids on the other. Sequences such as these that contain both hydrophobic and hydrophilic regions are said to be amphipathic. Mutations that disrupt the amphipathic character of these sequences usually disrupt targeting to the matrix, although many other amino acid substitutions do not. These findings indicate that the amphipathicity of matrix-targeting sequences is critical to their function.
The cell-free assay outlined in Figure 13-23 has been widely used to define the biochemical steps in the import of mitochondrial precursor proteins. Respiring (energized) mitochondria extracted from cells can incorporate mitochondrial precursor proteins that have been synthesized in the absence of mitochondria if they are carrying appropriate targeting sequences. Successful incorporation of the precursor into the organelle can be assayed by resistance to digestion by an added protease such as trypsin. In other assays, successful import of a precursor protein can be shown by the proper cleavage of the N-terminal targeting sequences by specific mitochondrial proteases. The uptake of completely presynthesized mitochondrial precursor proteins by the organelle in this system contrasts with the cell-free cotranslational translocation of secretory proteins into the ER, which generally occurs only when microsomal (ER-derived) membranes are present during synthesis (see Figure 13-4).
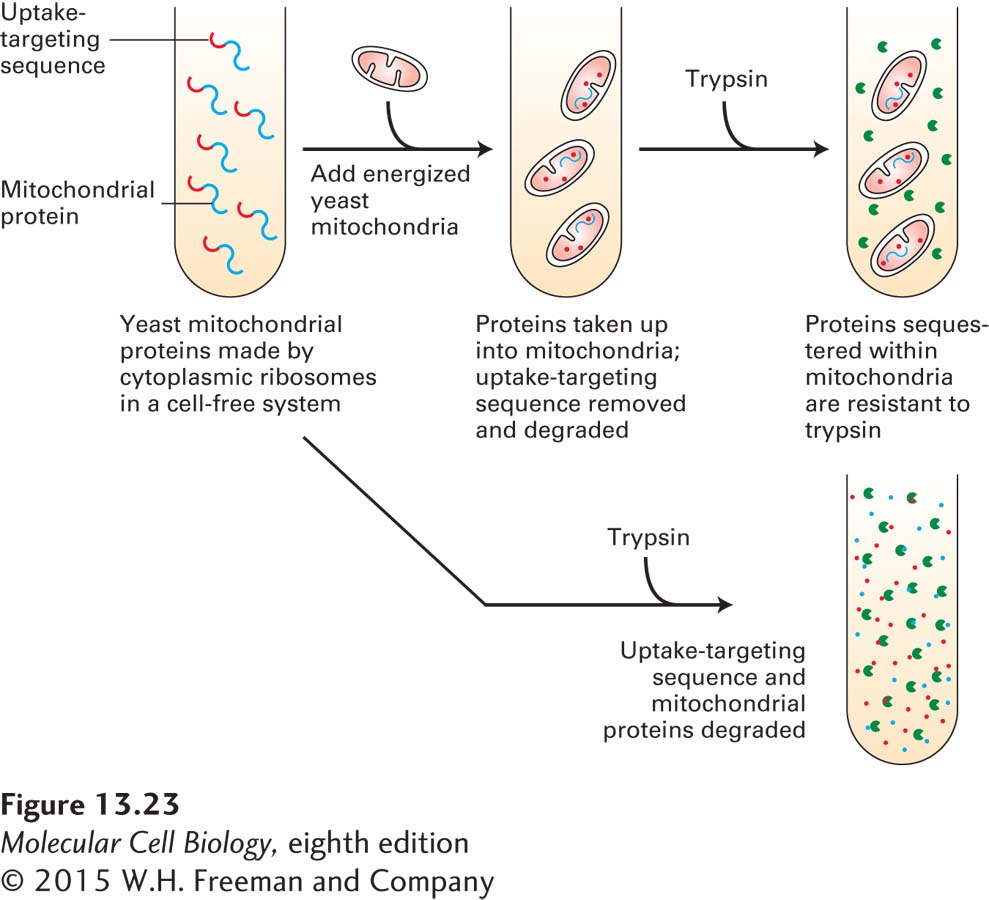
FIGURE 13-23 Import of mitochondrial precursor proteins is assayed in a cell-free system. Mitochondrial precursor proteins with attached uptake-targeting signals can be synthesized on ribosomes in a cell-free reaction. When respiring mitochondria are added to these synthesized mitochondrial precursor proteins (top), the proteins are taken up by mitochondria. Inside mitochondria, the proteins are protected from the action of proteases such as trypsin. When no mitochondria are present (bottom), the mitochondrial proteins are degraded by added protease. Only energized (respiring) mitochondria, which have a proton electrochemical gradient (proton-motive force) across the inner membrane, take up these proteins. Furthermore, the imported proteins must contain an appropriate uptake-targeting sequence. Uptake also requires ATP and a cytosolic extract containing chaperone proteins that maintain the precursor proteins in an unfolded conformation. This assay has been used to study targeting sequences and other features of the translocation process.
