Multiple Signals and Pathways Target Proteins to Submitochondrial Compartments
Unlike targeting to the matrix, targeting of proteins to the intermembrane space, inner membrane, or outer membrane of mitochondria generally requires more than one targeting sequence and occurs via one of several pathways. Figure 13-26 summarizes the organization of targeting sequences in proteins sorted to different mitochondrial locations.
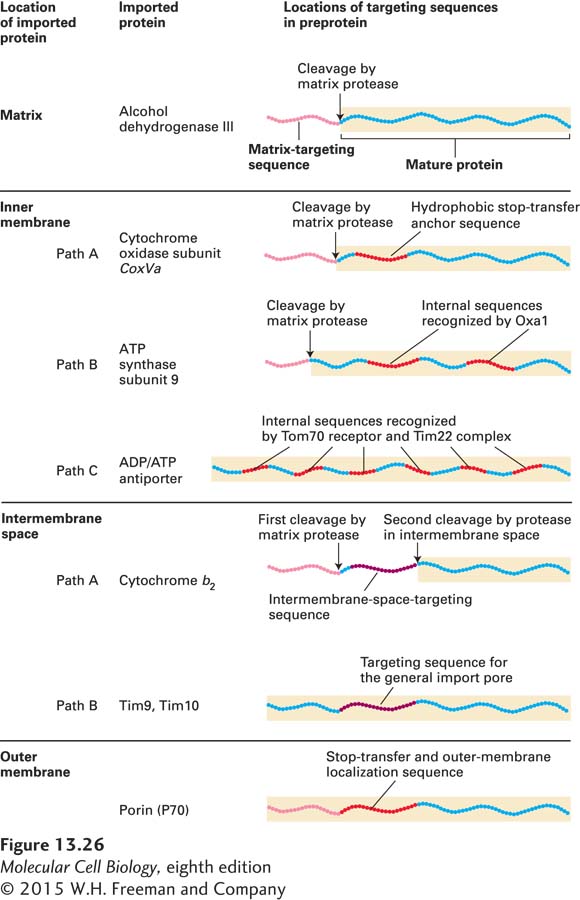
FIGURE 13-26 Targeting sequences in imported mitochondrial proteins. Most mitochondrial proteins have an N-terminal matrix-targeting sequence (pink) that is similar, though not identical, in different proteins. Proteins destined for the inner membrane, the intermembrane space, or the outer membrane have one or more additional targeting sequences that function to direct the proteins to these locations by several different pathways. The lettered pathways correspond to those illustrated in Figures 13-27 and 13-28. See W. Neupert, 1997, Annu. Rev. Biochem. 66:863.
Inner-Membrane Proteins Three separate pathways are known to target proteins to the inner mitochondrial membrane. One pathway makes use of the same machinery that is used for the targeting of matrix proteins (Figure 13-27, path A). A cytochrome oxidase subunit called CoxVa is one protein transported by this pathway. The precursor form of CoxVa, which contains an N-terminal matrix-targeting sequence recognized by the Tom20/22 import receptor, is transferred through the Tom40 general import pore of the outer membrane and the inner-membrane Tim23/17 translocation complex. In addition to the matrix-targeting sequence, which is cleaved during import, CoxVa contains a hydrophobic stop-transfer anchor sequence. As the protein passes through the Tim23/17 channel, the stop-transfer anchor sequence blocks translocation of the C-terminus across the inner membrane. The membrane-anchored intermediate is then transferred laterally into the bilayer of the inner membrane, much as type I integral membrane proteins are incorporated into the ER membrane (see Figure 13-11).
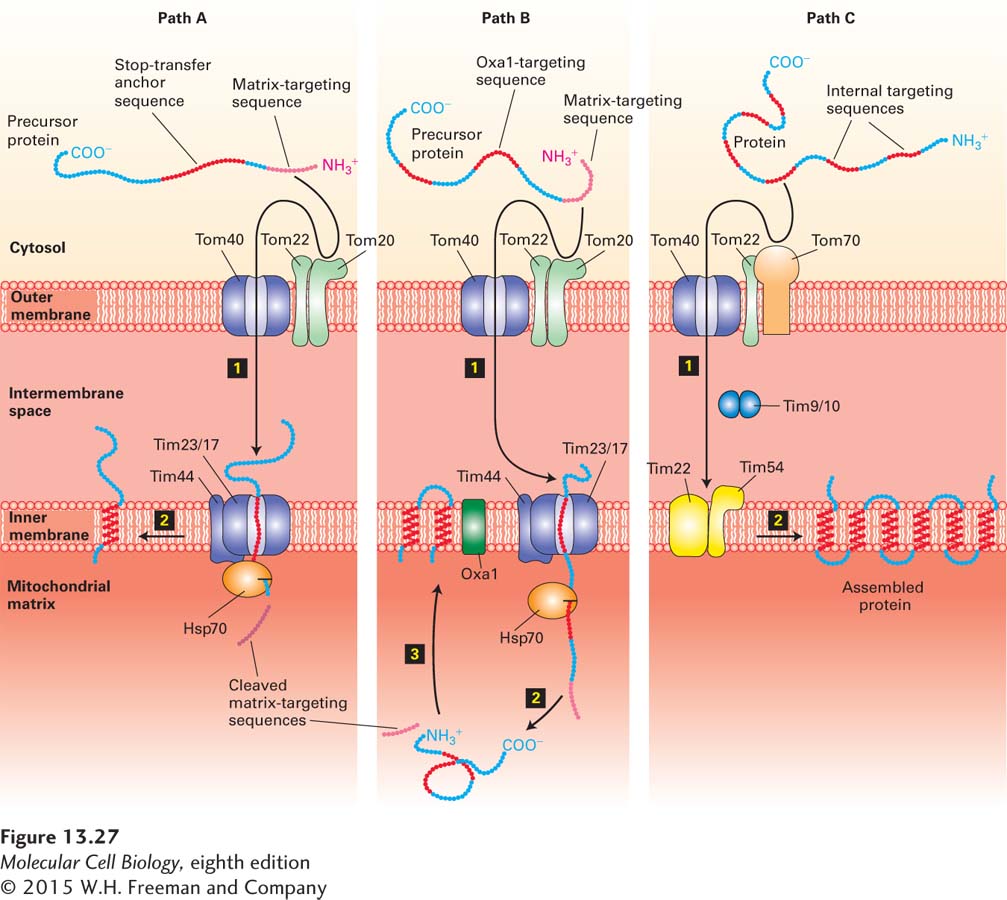
FIGURE 13-27 Three pathways to the inner mitochondrial membrane from the cytosol. Proteins with different targeting sequences are directed to the inner membrane via different pathways. In all three pathways, proteins cross the outer membrane via the Tom40 general import pore. Proteins delivered by pathways A and B contain an N-terminal matrix-targeting sequence that is recognized by the Tom20/22 import receptor in the outer membrane. Although both these pathways use the Tim23/17 inner-membrane channel, they differ in that the entire precursor protein enters the matrix and is then redirected to the inner membrane in pathway B. Matrix Hsp70 plays a role similar to its role in the import of soluble matrix proteins (see Figure 13-23). Proteins delivered by pathway C contain internal sequences that are recognized by the Tom70/Tom22 import receptor; a different inner-membrane translocation channel (Tim22/54) is used in this pathway. Two intermembrane proteins (Tim9 and Tim10) facilitate transfer between the outer and inner channels. See the text for discussion. See R. E. Dalbey and A. Kuhn, 2000, Annu. Rev. Cell Dev. Biol. 16:51, and N. Pfanner and A. Geissler, 2001, Nat. Rev. Mol. Cell Biol. 2:339.
A second pathway to the inner membrane is followed by proteins (e.g., ATP synthase subunit 9) whose precursors contain both a matrix-targeting sequence and internal hydrophobic domains recognized by an inner-membrane protein termed Oxa1. This pathway is thought to involve translocation of at least a portion of the precursor into the matrix via the Tom40 and Tim23/17 channels. After cleavage of the matrix-targeting sequence, the protein is inserted into the inner membrane by a process that requires interaction with Oxa1 and perhaps other inner-membrane proteins (Figure 13-27, path B). Oxa1 is related to a protein involved in inserting some plasma-membrane proteins in bacteria. This relatedness suggests that Oxa1 may have descended from the translocation machinery in the endosymbiotic bacterium that eventually became the mitochondrion. However, the proteins that form the inner-membrane channels in mitochondria are not related to the proteins in bacterial translocons. Oxa1 also participates in the inner-membrane insertion of certain proteins (e.g., subunit II of cytochrome oxidase) that are encoded by mitochondrial DNA and synthesized in the matrix by mitochondrial ribosomes.
The final pathway for insertion in the inner mitochondrial membrane is followed by multipass proteins that contain six membrane-spanning domains, such as the ATP/ADP antiporter. These proteins, which lack the usual N-terminal matrix-targeting sequence, contain multiple internal mitochondrial targeting sequences. After the internal sequences are recognized by a second import receptor composed of outer-membrane proteins Tom70 and Tom22, the imported protein passes through the outer membrane via the general import pore (Figure 13-27, path C). The protein then is transferred to a second translocation complex in the inner membrane composed of the Tim22, Tim18, and Tim54 proteins. Transfer to the Tim22/18/54 complex depends on a multimeric complex of two small proteins, Tim9 and Tim10, which reside in the intermembrane space. These small Tim proteins are thought to act as chaperones, guiding imported protein precursors from the general import pore to the Tim22/18/54 complex in the inner membrane by binding to their hydrophobic regions, preventing them from forming insoluble aggregates in the aqueous environment of the intermembrane space. Ultimately the Tim22/18/54 complex is responsible for incorporating the multiple hydrophobic segments of the imported protein into the inner membrane.
Intermembrane-Space Proteins Two pathways deliver cytosolic proteins to the space between the inner and outer mitochondrial membranes. The major pathway is followed by proteins, such as cytochrome b2, whose precursors carry two different N-terminal targeting sequences, both of which are ultimately cleaved. The most N-terminal of the two sequences is a matrix-targeting sequence, which is removed by the matrix protease. The second targeting sequence is a hydrophobic segment that blocks complete translocation of the protein across the inner membrane (Figure 13-28, path A). After the resulting membrane-embedded intermediate diffuses laterally away from the Tim23/17 translocation channel, a protease in the membrane cleaves the protein near the hydrophobic transmembrane segment, releasing the mature protein in a soluble form into the intermembrane space. Except for the second proteolytic cleavage, this pathway is similar to that of inner-membrane proteins such as CoxVa (see Figure 13-27, path A).
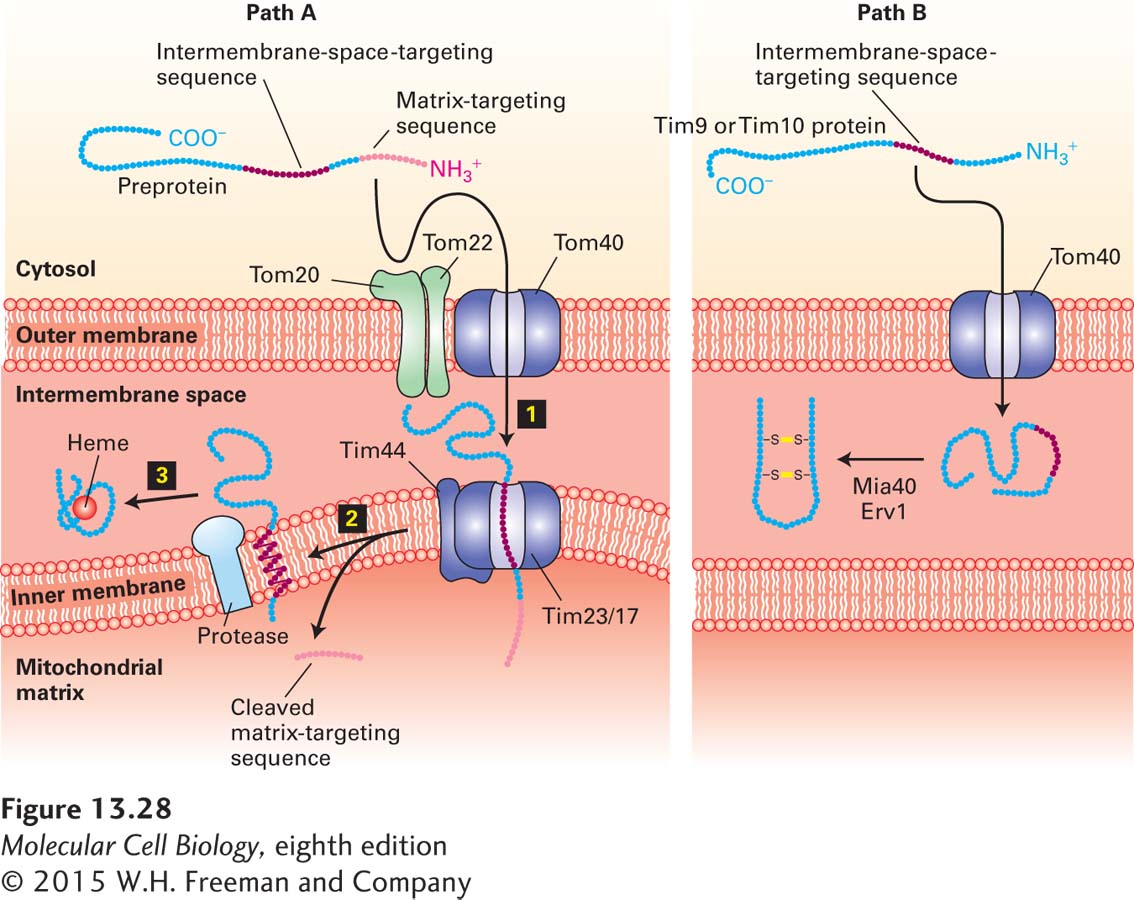
FIGURE 13-28 Two pathways to the mitochondrial intermembrane space. Pathway A, the major one for delivery of proteins from the cytosol to the intermembrane space, is similar to pathway A for delivery to the inner membrane (see Figure 13-26). The major difference is that the internal targeting sequence in these proteins, such as cytochrome b2, is recognized by an inner-membrane protease, which cleaves the protein on the intermembrane-space side of the membrane. The released protein then folds and binds to its heme cofactor within the intermembrane space. Pathway B is a specialized pathway for delivery of the proteins Tim9 and Tim10 to the intermembrane space. These proteins readily pass through the Tom40 general import pore, and once they are in the intermembrane space, they fold and form disulfide bonds that prevent reverse translocation through Tom40. The disulfide bonds are generated by Erv1 and are transferred to Tim9 and Tim10 by Mia40. See R. E. Dalbey and A. Kuhn, 2000, Annu. Rev. Cell Dev. Biol. 16:51; N. Pfanner and A. Geissler, 2001, Nat. Rev. Mol. Cell Biol. 2:339; and K. Tokatlidis, 2005, Cell 121:965–967.
The small Tim9 and Tim10 proteins, which reside in the intermembrane space, illustrate a second pathway for targeting to the intermembrane space. Proteins imported by this pathway do not contain an N-terminal matrix-targeting sequence and are delivered directly to the intermembrane space via the general import pore without involvement of any inner-membrane translocation factors (Figure 13-28, path B). Translocation through the Tom40 general import pore does not seem to be coupled to any energetically favorable process; however, once located in the intermembrane space, Tim9 and Tim10 proteins acquire two disulfide bonds each and fold into compact, stable structures. Apparently, the mechanism that drives their unidirectional translocation through the outer membrane involves passive diffusion through the outer membrane followed by folding and disulfide bond formation, which irreversibly traps the proteins in the intermembrane space. In many respects, the process of disulfide bond formation in the intermembrane space resembles that in the ER lumen and involves a disulfide bond–generating protein, Erv1, and a disulfide transfer protein, Mia40.
Outer-Membrane Proteins Many of the proteins that reside in the mitochondrial outer membrane, including the Tom40 pore itself and mitochondrial porin, have a β-barrel structure in which antiparallel β strands form hydrophobic transmembrane segments surrounding a central channel. Such proteins are incorporated into the outer membrane by first interacting with the general import pore, Tom40; they are then transferred to the SAM (sorting and assembly machinery) complex, which is composed of at least three outer-membrane proteins. One of these three proteins, Sam50, is closely related to the bacterial protein BamA, which is necessary for insertion of β-barrel proteins into the outer membrane of gram-negative bacteria. This relatedness provides another example of the conservation of the mechanism of membrane protein insertion between bacteria and mitochondria. Presumably it is the very stable hydrophobic nature of β-barrel proteins that ultimately causes them to be stably incorporated into the outer membrane, but precisely how the SAM complex facilitates this process is not known.


