On Binding Delta, the Notch Receptor Is Cleaved, Releasing a Component Transcription Factor
Both the receptor called Notch and its ligand Delta are single-spanning transmembrane proteins found on the cell surface. Notch also has other families of ligands, but the molecular mechanisms of activation are the same with each ligand. The extracellular domain of Delta on the signaling cell binds to Notch on an adjacent responding cell (but not on the same cell), activating Notch so that it undergoes two cleavage events; these events result in release of the Notch cytosolic domain, which functions as a transcription factor. Notch protein is synthesized as a monomeric membrane protein in the endoplasmic reticulum. In the Golgi complex, it undergoes a proteolytic cleavage that generates an extracellular subunit and a transmembrane-cytosolic subunit; the two subunits remain noncovalently associated with each other.
ADAM 10 is a matrix metalloprotease (MMP), a member of a class of metal-containing enzymes that cleave the extracellular segments of target proteins near the extracellular surface of the plasma membrane. ADAM 10 performs the initial cleavage of the Notch extracellular domain, but in the absence of Delta on an adjacent cell, the Notch extracellular domain is folded such that ADAM 10 cannot access the protease cleavage site (Figure 16-36).
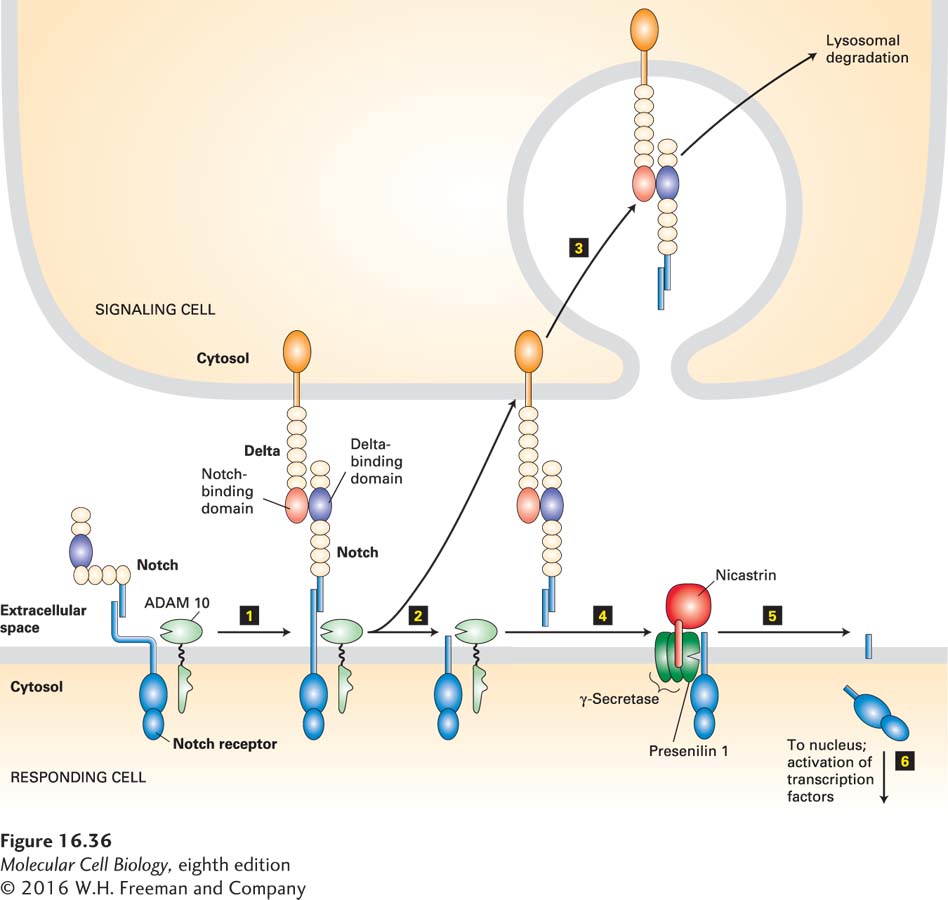
FIGURE 16-36 Notch/Delta signaling pathway. In the absence of Delta, the transmembrane subunit of Notch on a responding cell is noncovalently associated with its extracellular subunit; the extracellular domain is folded so that it cannot be cleaved by the cell-surface protease ADAM 10. Binding of Notch to its ligand Delta on an adjacent signaling cell (step 1) is followed by endocytosis of Delta by the signaling cell, stretching the Notch extracellular domain so that ADAM 10 can cleave it (step 2). The released Notch extracellular domain remains bound to Delta and is endocytosed by the signaling cell (step 3). Next the nicastrin subunit (colored red) of the four-protein γ-secretase complex binds to the stump generated by ADAM 10 (step 4), and then the protease, presenilin 1, catalyzes an intramembrane cleavage that releases the cytosolic segment of Notch (step 5). Following translocation to the nucleus, this Notch segment interacts with several transcription factors to affect expression of genes that in turn influence the determination of cell fate during development (step 6). See D. Seals and S. Courtneidge, 2003, Gene Dev. 17:7, and L. Meloty-Kapella et al., 2012, Dev. Cell 22:1299.
Following the binding of Delta to a Notch protein on the responding cell, Delta in the signaling cell undergoes endocytosis. The force accompanying the movement of Delta into the signaling cell stretches the Notch protein, changing its conformation and allowing access by ADAM 10, which cleaves off the Notch extracellular domain. The Notch extracellular domain remains bound to Delta, is internalized by the signaling cell, and is probably degraded in lysosomes.
The second cleavage of Notch occurs within the hydrophobic membrane-spanning region of Notch and is catalyzed by a four-protein transmembrane complex termed γ-secretase. This cleavage releases the Notch cytosolic segment, which immediately translocates to the nucleus, where it affects transcription of target genes. Its effect, like those of other transcription factors activated downstream of other cell-surface receptors, depends on the constellation of epigenetic chromatin marks and the presence of cell-specific transcription factors.
The four-protein γ-secretase complex contains presenilin 1 (PS1), the actual protease, and three other essential subunits, aph-1, pen-2, and nicastrin. How peptide bond hydrolysis can occur within a hydrophobic intramembrane environment is not well understood because the molecular structure of presenilin is not known to sufficient resolution (see Figure 16-37b). But the molecular structure of a related archaeal protein, also with nine transmembrane segments, suggests that two aspartate residues, located near each other on the cytosolic side of two transmembrane helices, are essential for catalysis and are surrounded by water; thus proteolytic cleavage probably occurs in a partially aqueous environment.
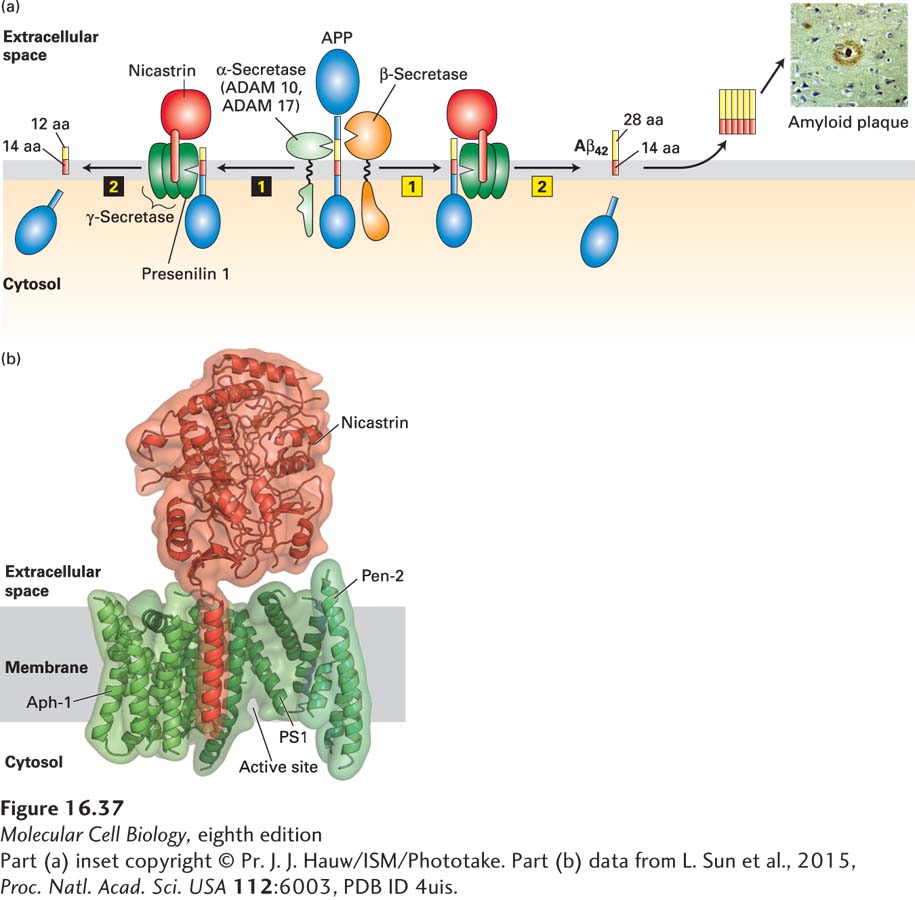
FIGURE 16-37 Proteolytic cleavage of APP and Alzheimer’s disease. (a, left) Sequential proteolytic cleavage by α-secretase (ADAM 10 or ADAM 17) (step 1) and γ-secretase (step 2) produces an innocuous membrane-embedded peptide of 26 amino acids. (a, right) Cleavage in the extracellular domain by β-secretase (step 1) followed by cleavage within the membrane by γ-secretase (step 2) generates the 42-amino-acid Aβ42 peptide, which spontaneously forms oligomers, and then the large amyloid plaques found in the brains of patients with Alzheimer’s disease (inset). In both pathways, the cytosolic segment of APP is released into the cytosol, but its function is not known. See S. Lichtenthaler and C. Haass, 2004, J. Clin. Invest. 113:1384, and V. Wilquet and B. De Strooper, 2004, Curr. Opin. Neurobiol. 14:582. (b) Three-dimensional structure of human γ-secretase at 0.45 nm resolution. It contains a total of 19 transmembrane segments and a large extracellular domain from nicastrin. The protease catalytic site in PS1 is located near the cytosolic surface. See P. Lu et al., 2014, Nature 512:166.
[Part (a) inset copyright © Pr. J. J. Hauw/ISM/Phototake. Part (b) data from L. Sun et al., 2015, Proc. Natl. Acad. Sci. USA 112:6003, PDB ID 4uis.]
Studies on cells and mice lacking nicastrin revealed why γ-secretase can cleave only proteins that have first been cleaved by an ADAM or other matrix metalloprotease. Nicastrin binds to the N-terminal extracellular stump of the membrane protein that is generated by the first protease (Figure 16-36, step 4). Without this stump, nicastrin, and thus the entire γ-secretase complex, cannot interact with its target protein. We examine the role of ADAM proteins and γ-secretase in the development of Alzheimer’s disease below.

