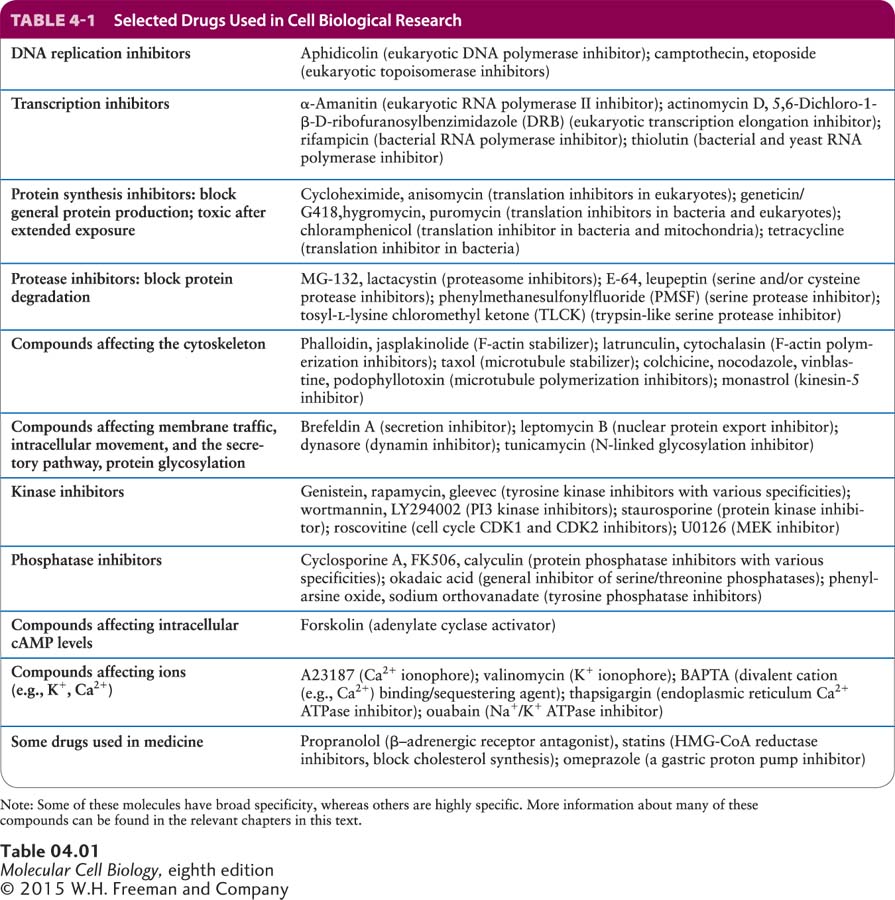
Drugs Are Commonly Used in Cell Biological Research
Another powerful way to analyze biological processes is to treat cells with drugs that bind to specific cell components and inactive or activate them. In this section, we discuss some of the common drugs used for this purpose and how new drugs affecting specific cell processes can be developed.
Naturally occurring drugs have been used for centuries, but how they worked was often unknown. For example, extracts of meadow saffron were used to treat gout, a painful disease resulting from inflammation of joints. Today we know that this plant contains colchicine, a drug that depolymerizes microtubules and interferes with the ability of white blood cells to move to sites of inflammation (see Chapter 18). Alexander Fleming discovered that certain fungi secrete compounds that kill bacteria (antibiotics), and his discovery resulted in the development of penicillin. Only later was it was discovered that penicillin inhibits cell division by blocking the assembly of the cell walls of certain bacteria.
Discoveries like these have resulted in a wide range of drugs that can inhibit specific and essential processes of cells. In most cases, researchers have eventually been able to identify the molecular targets of these drugs. For example, there are many antibiotic drugs that affect aspects of prokaryotic protein synthesis. A selection of some of the drugs most commonly used in cell biological research are listed in Table 4-1, grouped according to the process they inhibit.
How does one discover a new drug? One widely used approach is to search chemical libraries, consisting of tens of thousands to hundreds of thousands of different compounds, for chemicals that inhibit a specific process. The screening of chemical libraries in conjunction with high-throughput microscopic techniques has now become one of the major routes for new leads in drug discovery. Here we give just one case to illustrate how this type of approach works.
In our example (Figure 4-7a), researchers wanted to identify compounds that inhibit mitosis, the process by which duplicated chromosomes are accurately segregated by a microtubule-based machine called the mitotic spindle (discussed in Chapter 18). It was known that if spindle assembly is compromised, cells are arrested in mitosis. Therefore, the screen first used an automated robotic method to look for compounds that arrest cells in mitosis. The basis for the inhibition of mitosis by the candidate compounds was then explored to see if they affected assembly of the microtubules. Since inhibition of microtubule assembly was not of interest, the effect of the remaining candidate compounds on the structure of the spindle was determined by immunofluorescence microscopy using antibodies to tubulin, the major protein of microtubules. Over 16,000 compounds were screened, and a compound was identified that resulted in cells with abnormal spindles—instead of having two asters, they had a single aster, resulting in what is called a mono-astral array (Figure 4-7b). This drug, now called monastrol, was found to interfere with the assembly of the spindle by inhibiting a microtubule-based motor protein called kinesin-5. (See Chapter 18 for more details about the mitotic spindle.) Derivatives of monastrol are now being tested as anti-tumor agents for the treatment of certain cancers.
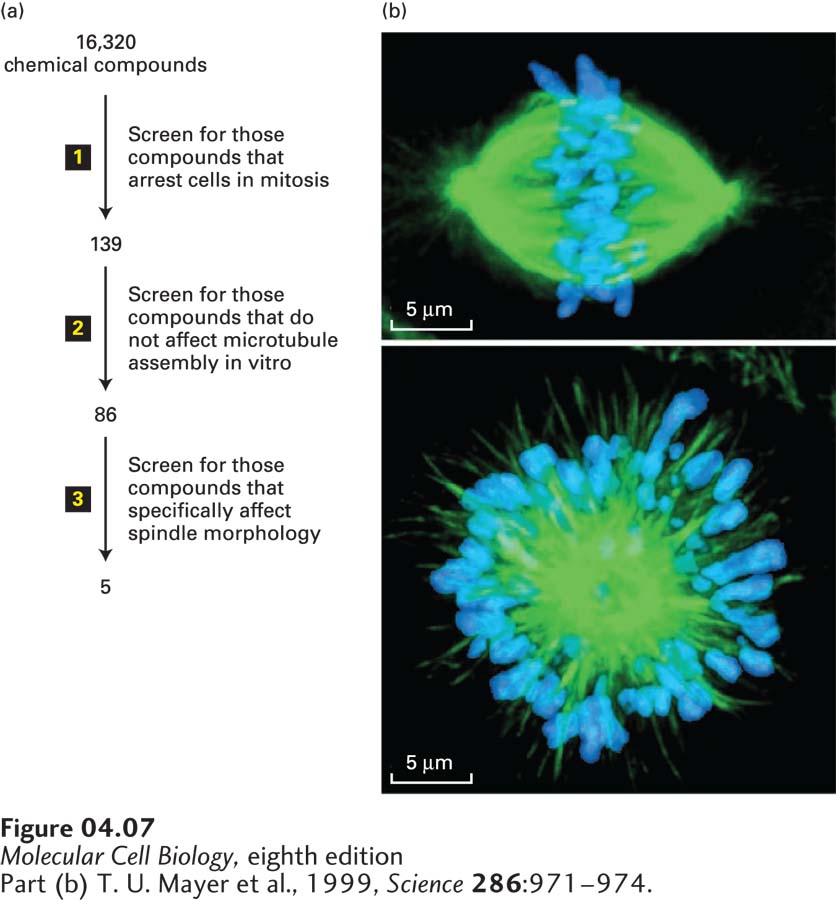
FIGURE 4-7 Screening for drugs that affect specific biological processes. (a) In this example, a chemical library of 16,320 different chemicals was subjected to a series of screens for inhibitors of mitosis. Since such an inhibitor is expected to arrest cells at the mitotic stage of the cell cycle, the first screen (step 1) was to see if any of the chemicals enhanced the level of a marker specific for mitotic cells; this screen yielded 139 candidates. Microtubules make up the structure of the mitotic spindle, and the researchers were not interested in new drugs that target microtubules, so in the second screen (step 2) they tested the 139 compounds for their ability to affect microtubule assembly; this test eliminated 53 candidates. Immunofluorescence microscopy with antibodies to tubulin (the major subunit of microtubules), together with a stain for DNA, was then used in the third screen (step 3) to identify compounds that disrupt the structure of the spindle. (b) Localization of tubulin (green) and DNA (blue) for an untreated mitotic spindle (top) and one treated with one of the recovered compounds, now called monastrol. Monastrol inhibits a microtubule-based motor protein called kinesin-5, discussed in Chapter 18, that is necessary to separate the poles of the mitotic spindle. When kinesin-5 is inhibited, the two poles remain associated to give a monopolar spindle.
[Part (b) T. U. Mayer et al., 1999, Science 286:971–974.]