Polysomes and Rapid Ribosome Recycling Increase the Efficiency of Translation
Translation of a single eukaryotic mRNA molecule to yield a typical-sized protein takes 1 to 2 minutes. Two phenomena significantly increase the overall rate at which cells can synthesize a protein: the simultaneous translation of a single mRNA molecule by multiple ribosomes, and rapid recycling of ribosomal subunits after they disengage from a stop codon. Simultaneous translation of an mRNA by multiple ribosomes is readily observable in electron micrographs and by sedimentation analysis, revealing mRNA molecules attached to multiple ribosomes bearing nascent growing polypeptide chains. These structures, referred to as polyribosomes or polysomes, were seen to be circular in electron micrographs of some tissues. Subsequent studies with purified initiation factors explained the circular shape of polyribosomes and suggested the mechanism by which ribosomes recycle efficiently.
These studies revealed that multiple copies of the cytoplasmic poly(A)-binding protein (PABPC) interact with both an mRNA poly(A) tail and the eIF4G subunit of eIF4. Since the eIF4E subunit of eIF4 binds to the cap structure on the 5′ end of an mRNA, the two ends of an mRNA molecule are bridged by the intervening proteins, forming a “circular” mRNA (Figure 5-27a). Because the two ends of a polysome are relatively close together, ribosomal subunits that disengage from a stop codon are positioned near the 5′ end, facilitating reinitiation by the interaction of the 40S subunit and its associated initiation factors with eIF4 bound to the 5′ cap. The circular pathway depicted in Figure 5-27b is thought to enhance ribosome recycling and thus increase the efficiency of protein synthesis.
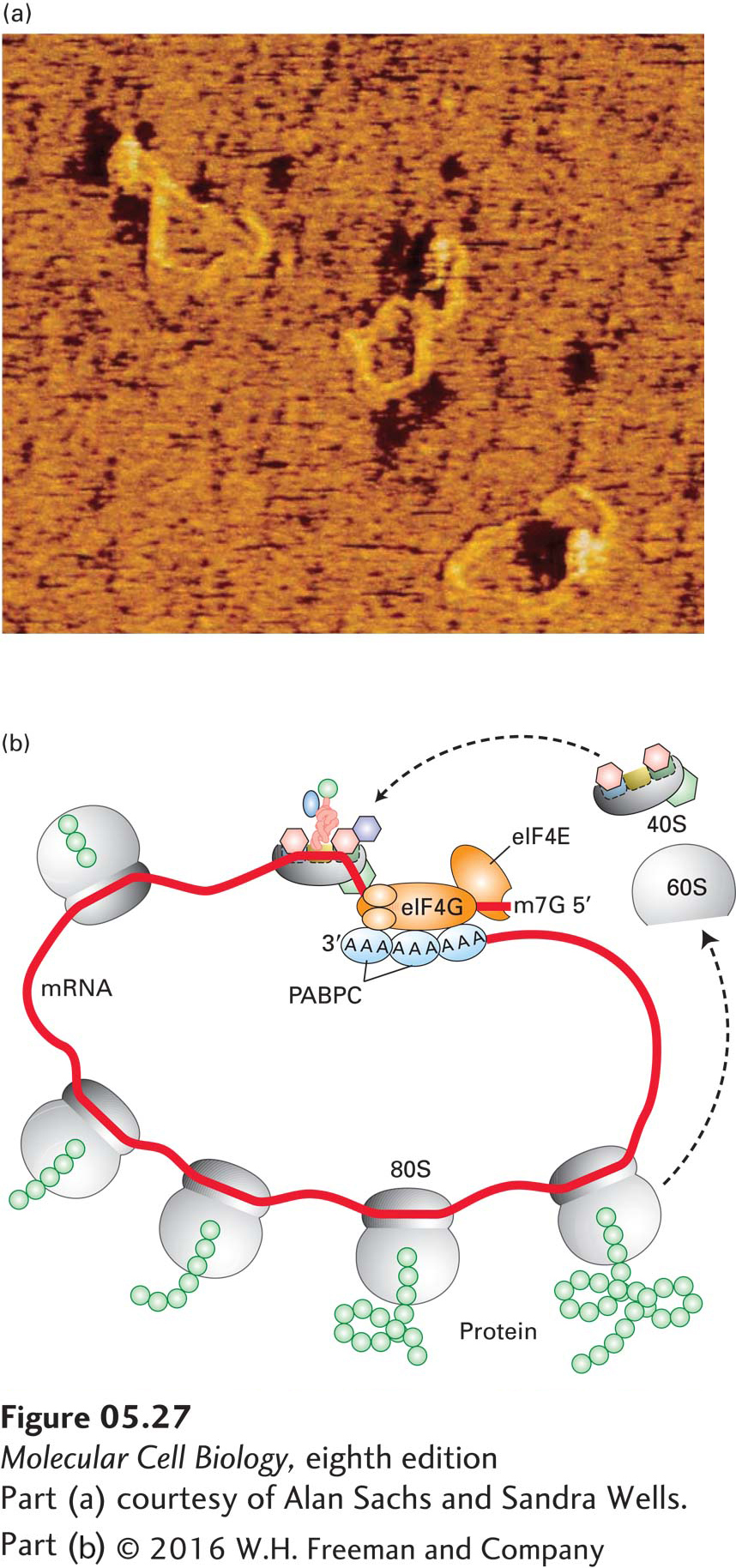
EXPERIMENTAL FIGURE 5-27 Circular structure of mRNA increases translation efficiency. Eukaryotic mRNA forms a circular structure owing to interactions of three proteins. (a) In the presence of purified yeast poly(A)-binding protein [PABP; there is only one PABP in S. cerevisiae, rather than a nuclear (PABPN) and cytoplasmic (PABPC) protein as in higher eukaryotes], eIF4E, and eIF4G, eukaryotic mRNAs form circular structures, visible in this force-field electron micrograph. In these structures, protein-protein and protein-mRNA interactions form a bridge between the 5′ and 3′ ends of the mRNA. (b) Model of protein synthesis on circular polysomes and recycling of ribosomal subunits. Multiple individual ribosomes can simultaneously translate a eukaryotic mRNA, shown here in a circular form stabilized by interactions between proteins bound at the 3′ and 5′ ends. When a ribosome completes translation and dissociates from the 3′ end, the separated subunits can rapidly find the nearby 5′ cap (m7G) and PABPC-bound poly(A) tail and initiate another round of synthesis.
[Part (a) courtesy of Alan Sachs and Sandra Wells.]
