Several Proteins Participate in DNA Replication
Detailed understanding of the eukaryotic proteins that participate in DNA replication initially came largely from studies with small viral DNAs, particularly SV40 DNA, the circular genome of a virus that infects monkeys. Virus-infected cells replicate large numbers of the simple viral genome in a short period of time, which makes them an ideal model system for studying basic aspects of DNA replication. Because simple viruses such as SV40 depend largely on the DNA replication machinery of their host cells (in this case monkey cells), they offer a unique opportunity to study the replication of multiple identical small DNA molecules by cellular proteins. Figure 5-30 depicts the multiple proteins that coordinate the copying of SV40 DNA at a replication fork. The assembled proteins at a replication fork further illustrate the concept of molecular machines introduced in Chapter 3. These multicomponent complexes permit the cell to carry out an ordered sequence of events that accomplishes essential cell functions.
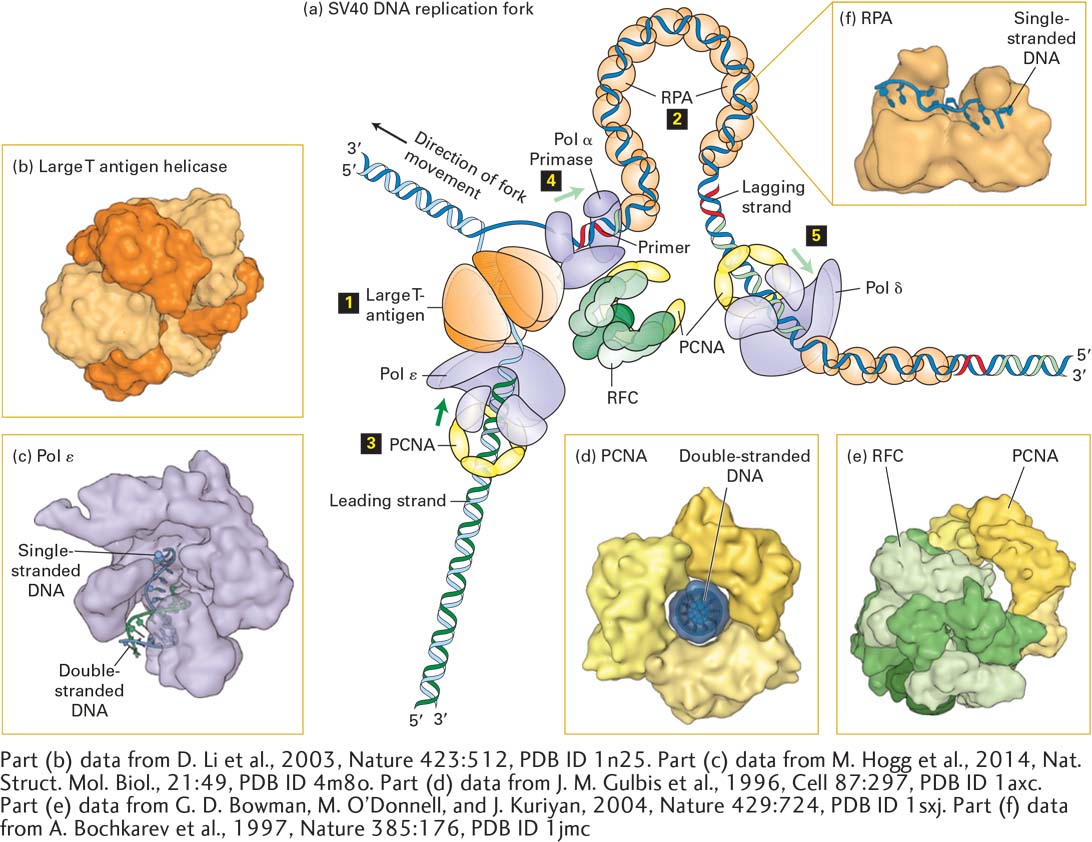
FIGURE 5-30 Model of an SV40 DNA replication fork. (a) A hexamer of large T-antigen, a viral protein, functions as a helicase to unwind the parent DNA strands. The leading strand is extended by DNA polymerase ε (Pol ε) up to the replication fork. Pol ε is bound to a ring of PCNA that surrounds the daughter double-stranded DNA so that the Pol ε-PCNA complex remains stably associated with the replication fork. The single-stranded region of the lagging strand template generated by T-antigen helicase is bound by multiple copies of the heterotrimeric protein RPA. Primers for lagging-strand synthesis (red, RNA; light green, DNA) are synthesized by a complex of primase and DNA polymerase α (Pol α). The 3′ end of each primer synthesized by Pol α–primase is then bound by a PCNA–Pol δ complex, which extends the primer and synthesizes most of each Okazaki fragment. (b) The helicase domain of SV40 T-antigen forms a hexameric replicative helicase. Subunits are shown in alternating light and dark orange. (c) Model of DNA polymerase ε extending the 3’-end of the leading strand. (d) The three subunits of PCNA, shown in different shades of yellow, form a circular structure with a central hole through which daughter double-stranded DNA passes. (e) RFC, the pentameric “clamp-loader” (monomers shown in different shades of green) is shown bound to a circular trimer PCNA before the PCNA “clamp” is opened. (f) The large subunit of RPA contains two domains that bind single-stranded DNA. Note that the single DNA strand is extended, with the bases oriented in an optimal conformation for replication by Pol δ. See M. O’Donnell, L. Langston, and B. Stillman, 2013, Cold Spring Harbor Perspect. Biol. 5:a010108.
[Part (b) data from D. Li et al., 2003, Nature 423:512, PDB ID 1n25. Part (c) data from M. Hogg et al., 2014, Nat. Struct. Mol. Biol., 21:49, PDB ID 4m8o. Part (d) data from J. M. Gulbis et al., 1996, Cell 87:297, PDB ID 1axc. Part (e) data from G. D. Bowman, M. O’Donnell, and J. Kuriyan, 2004, Nature 429:724, PDB ID 1sxj. Part (f) data from A. Bochkarev et al., 1997, Nature 385:176, PDB ID 1jmc.]
The molecular machine that replicates SV40 DNA contains only one viral protein; all other proteins involved in SV40 DNA replication are provided by the host cell. This viral protein, large T-antigen, forms a hexameric replicative helicase, a protein that uses energy from ATP hydrolysis to unwind the parent strands at a replication fork. Primers for the leading and lagging daughter strands are synthesized by a complex of primase, which synthesizes a short RNA primer (~12 nucleotides), and DNA polymerase α (Pol α), which extends the RNA primer with deoxyribonucleotides for another 25 nucleotides or so, forming a mixed RNA-DNA primer.
The primer is extended into daughter-strand DNA by DNA polymerase δ (Pol δ), which is less likely to make errors during copying of the template strand than is Pol α because of its proofreading mechanism (see Section 5.6). During the replication of cellular DNA, Pol δ synthesizes lagging-strand DNA, while DNA polymerase ε (Pol ε) synthesizes most of the length of the leading strand. Pol δ and Pol ε each form a complex with PCNA (proliferating cell nuclear antigen), which displaces the primase–Pol α complex following primer synthesis. As illustrated in Figure 5-30d, PCNA is a homotrimeric protein that has a central hole through which the daughter duplex DNA passes, thereby preventing the PCNA-Pol δ and PCNA-Pol ε complexes from dissociating from the template. As such, PCNA is known as a sliding clamp that enables Pol δ and Pol ε to remain stably associated with a single template strand for thousands of nucleotides. A pentameric protein called RFC (replication factor C) functions to open the PCNA ring so that it can encircle the short region of double-stranded DNA synthesized by Pol α. Consequently, RFC is called a clamp loader.
After parent DNA is separated into single-stranded templates at the replication fork, the leading strand is extended by Pol ε, which can extend the growing strand up to the replication fork. The single-stranded template for lagging-strand synthesis is bound by multiple copies of RPA (replication protein A), a heterotrimeric protein (Figure 5-30c). Binding of RPA maintains the template in a uniform conformation that is optimal for copying by Pol δ. Bound RPA proteins are dislodged from the parent strand by Pol δ as it synthesizes the complementary strand base-paired with the parent strand.
Several other eukaryotic proteins that function in DNA replication are not depicted in Figure 5-30. For example, topoisomerase I associates with the parent DNA ahead of the replicative helicase (i.e., to the left of large T-antigen in Figure 5-30) to remove torsional stress introduced by the unwinding of the parent strands (see Figure 5-8a). Ribonuclease H and FEN I remove the ribonucleotides at the 5′ ends of Okazaki fragments; these ribonucleotides are replaced by deoxyribonucleotides added by Pol δ as it extends the upstream Okazaki fragment. Successive Okazaki fragments are coupled by DNA ligase through standard 5′→3′ phosphoester bonds. Other specialized DNA polymerases are involved in the repair of mismatches and lesions in DNA (see Section 5.6).
