DNA Replication Occurs Bidirectionally from Each Origin
As indicated in Figures 5-29 and 5-30, both parent DNA strands that are exposed by local unwinding at a replication fork are copied into daughter strands. In theory, DNA replication from a single origin could involve one replication fork that moves in one direction. Alternatively, two replication forks might assemble at a single origin and then move in opposite directions, leading to bidirectional growth of both daughter strands. Several types of experiments, including the one shown in Figure 5-31, provided early evidence in support of bidirectional strand growth.
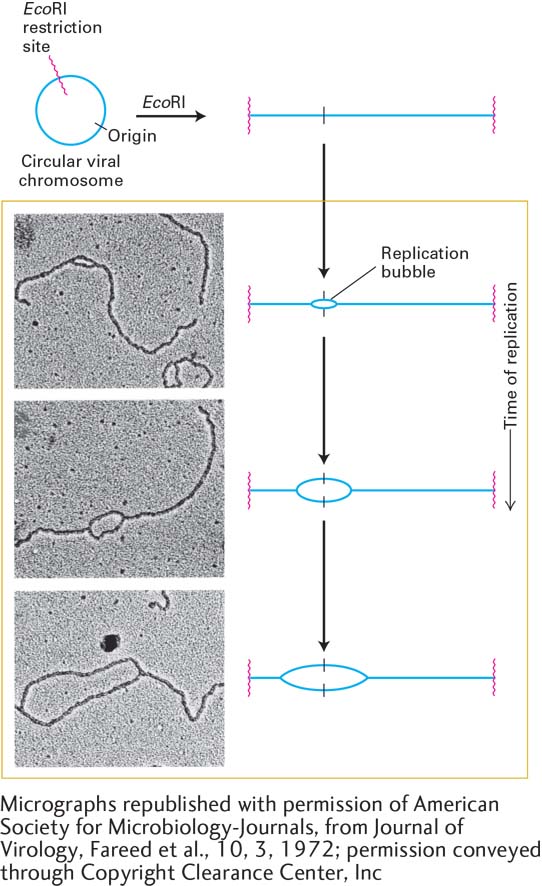
EXPERIMENTAL FIGURE 5-31 Bidirectional replication in SV40 DNA. Electron microscopy of replicating SV40 DNA indicates bidirectional growth of DNA strands from an origin. Replicating viral DNA from SV40-infected cells was cut by the restriction enzyme EcoRI, which recognizes one site in the circular viral DNA. This was done to provide a landmark in the SV40 genome: the EcoRI recognition sequence could now be easily recognized as the ends of the linear DNA molecules visualized by electron microscopy. Electron micrographs of EcoRI-cut, replicating SV40 DNA molecules showed a collection of cut molecules with increasingly longer replication “bubbles,” whose centers were a constant distance from each end of the cut molecules. This finding is consistent with chain growth in two directions from a common origin located at the center of a bubble, as illustrated in the corresponding diagrams. See G. C. Fareed et al., 1972, J. Virol. 10:484.
[Micrographs republished with permission of American Society for Microbiology-Journals, from Journal of Virology, Fareed et al., 10, 3, 1972; permission conveyed through Copyright Clearance Center, Inc.]
The general consensus is that all bacterial, archaeal, and eukaryotic cells employ a bidirectional mechanism of DNA replication. In the case of SV40 DNA, replication is initiated by the binding of two large T-antigen hexameric helicases to the single SV40 origin and the assembly of other proteins to form two replication forks. These forks then move away from the SV40 origin in opposite directions, and leading- and lagging-strand synthesis occurs at both forks. As shown in Figure 5-32, the left replication fork extends DNA synthesis in the leftward direction; similarly, the right replication fork extends DNA synthesis in the rightward direction.
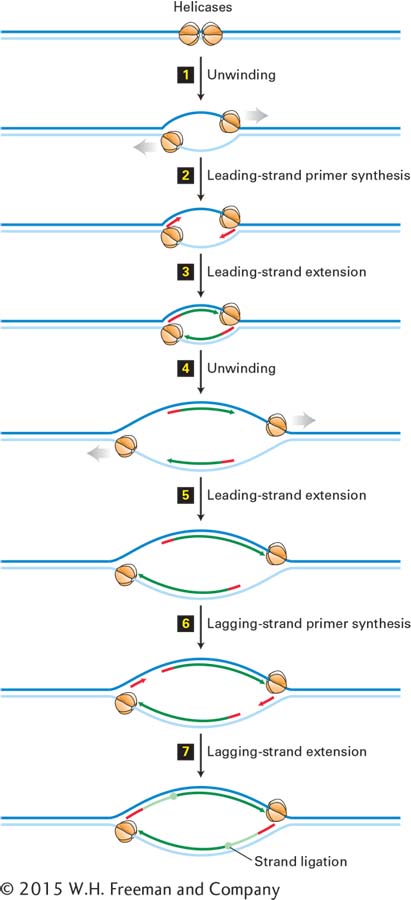
FIGURE 5-32 Bidirectional mechanism of DNA replication. The left replication fork here is comparable to the replication fork diagrammed in Figure 5-30 (although that figure also shows proteins other than large T-antigen, which are not shown here). Top: Two large T-antigen hexameric helicases first bind at the replication origin in opposite orientations. Step 1: Using energy provided by ATP hydrolysis, the helicases move in opposite directions, unwinding the parent DNA and generating single-stranded templates, which are bound by RPA proteins. Step 2: Primase–Pol α complexes synthesize short primers (red) base-paired to each of the separated parent strands. Step 3: PCNA-Rfc–Pol ε complexes replace the primase–Pol α complexes and extend the short primers, generating the leading strands (dark green) at each replication fork. Step 4: The helicases further unwind the parent strands, and RPA proteins bind to the newly exposed single-stranded regions. Step 5: PCNA-Rfc–Pol ε complexes extend the leading strands farther. Step 6: Primase–Pol α complexes synthesize primers for lagging-strand synthesis at each replication fork. Step 7: PCNA-Rfc–Pol δ complexes displace the primase–Pol α complexes and extend the lagging-strand Okazaki fragments (light green), which are eventually ligated to the 5′ ends of the leading strands. The position where ligation occurs is represented by a circle. Replication continues by further unwinding of the parent strands and synthesis of leading and lagging strands as in Steps 4–7. Although depicted as individual steps for clarity, unwinding and synthesis of leading and lagging strands occur concurrently.
Unlike SV40 DNA, eukaryotic chromosomal DNA molecules contain multiple replication origins separated by tens to hundreds of kilobases. A six-subunit protein called ORC, for origin recognition complex, binds to each origin and associates with other proteins required to load cellular hexameric helicases composed of six homologous MCM proteins. Two MCM helicases, oriented in opposite directions, separate the parent strands at an origin, and RPA proteins bind to the resulting single-stranded DNA. Synthesis of primers and subsequent steps in the replication of cellular DNA are thought to be analogous to those in SV40 DNA replication (see Figures 5-31 and 5-32).
Replication of cellular DNA and other events leading to the proliferation of cells must be tightly regulated so that the appropriate numbers of cells constituting each tissue will be produced during embryonic development and throughout the life of an organism. Control of the initiation step is the primary mechanism for regulating cellular DNA replication. Activation of MCM helicase, which is required to initiate cellular DNA replication, is regulated by a specific protein kinase (DDK), which in turn is regulated by S-phase cyclin-dependent kinases. Other cyclin-dependent kinases regulate additional aspects of cell proliferation, including the complex process of mitosis by which a eukaryotic cell divides into two daughter cells. Mitosis and another specialized type of cell division called meiosis, which generates haploid sperm and egg cells, are discussed in Chapter 6. We discuss the various regulatory mechanisms that determine the rate of cell division in Chapter 19.

