cDNA Libraries Represent the Sequences of Protein-Coding Genes
Genomic libraries are ideal for representing the genetic content of relatively simple organisms such as bacteria or yeast, but present certain experimental difficulties for higher eukaryotes. First, the genes of eukaryotes usually contain extensive intron sequences and can therefore be too large to be inserted intact into plasmid vectors. As a result, the sequences of individual genes are broken apart and carried in more than one clone. Moreover, the presence of introns and long intergenic regions in genomic DNA often makes it difficult to identify the important parts of a gene that actually encode protein sequences. For example, only about 1.5 percent of the human genome actually represents protein-coding gene sequences. Thus for many studies, cellular mRNAs, which lack the noncoding regions present in genomic DNA, are a more useful starting material for generating a DNA library. In this approach, DNA copies of mRNAs, called complementary DNAs (cDNAs), are synthesized and cloned into plasmid vectors. A large collection of the resulting cDNA clones, representing all the mRNAs expressed in a cell type, is called a cDNA library.
The first step in preparing a cDNA library is to isolate the total mRNA from the cell type or tissue of interest. Because of their poly(A) tails, mRNAs are easily separated from the much more prevalent rRNAs and tRNAs present in a cell extract by use of a matrix to which short strings of thymidylate (oligo-dTs) are linked. The general procedure for preparing a cDNA library from a mixture of cellular mRNAs is outlined in Figure 6-17. The enzyme reverse transcriptase, which is found in retroviruses, is used to synthesize a strand of DNA complementary to each mRNA molecule, starting from an oligo-dT primer (steps 1 and 2 ). The resulting cDNA-mRNA hybrid molecules are converted in several steps into double-stranded cDNA molecules corresponding to all the mRNA molecules in the original preparation (steps 3 –5). Each double-stranded cDNA contains an oligo-dC⋅oligo-dG double-stranded region at one end and an oligo-dT⋅oligo-dA double-stranded region at the other end. Methylation of the cDNA protects it from subsequent restriction enzyme cleavage (step 6).
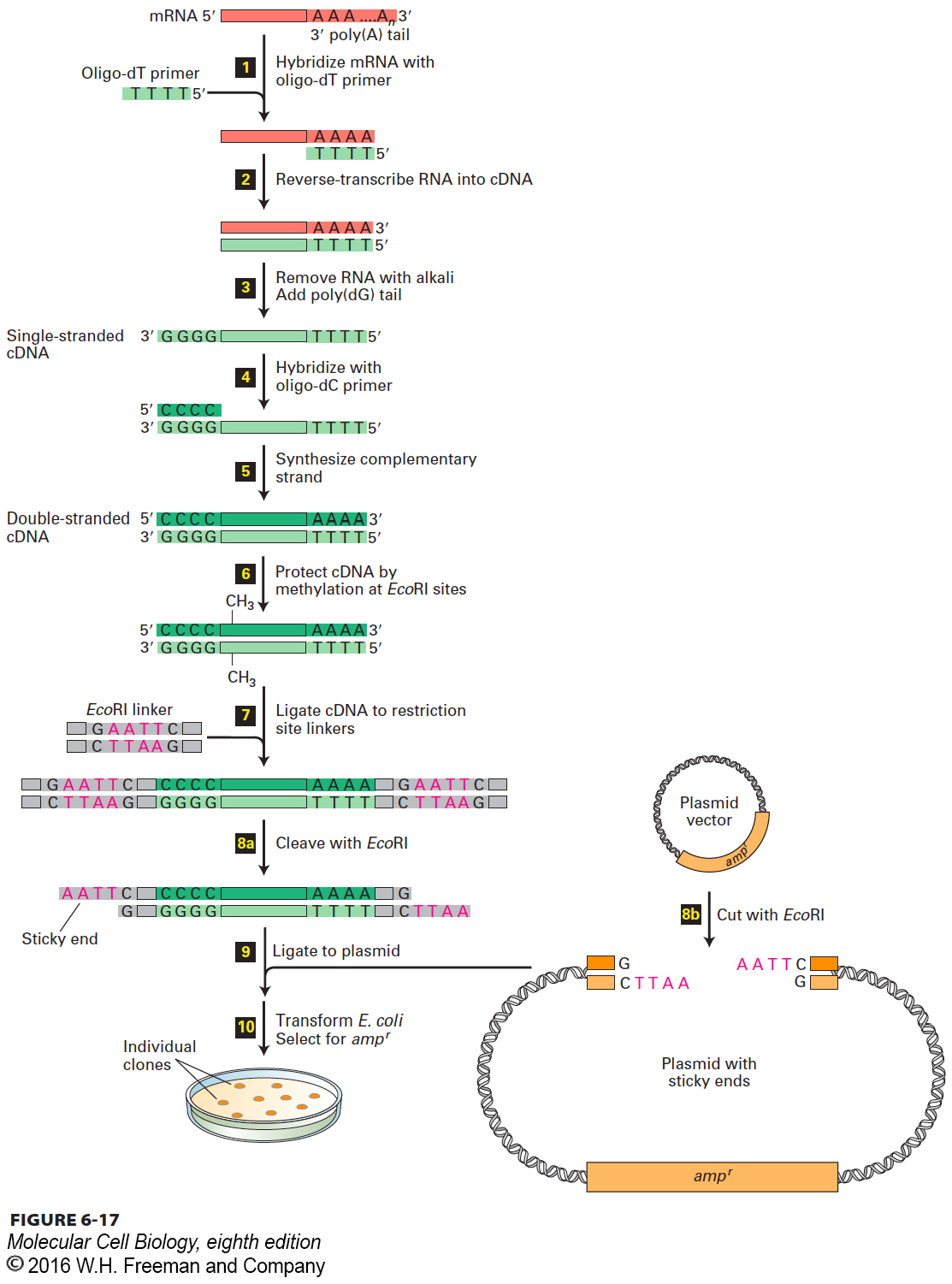
FIGURE 6-17 A cDNA library contains representative copies of cellular mRNA sequences. A mixture of mRNAs is the starting point for preparing recombinant plasmid clones, each containing a cDNA. Transforming E. coli with the recombinant plasmids generates a set of cDNA clones representing all the cellular mRNAs. See the text for a step-by-step discussion.
To prepare double-stranded cDNAs for cloning, short double-stranded DNA molecules containing the recognition site for a particular restriction enzyme (called linkers) are ligated to both ends of the cDNAs using DNA ligase from bacteriophage T4 (Figure 6-17, step 7). As noted earlier, this ligase can join “blunt-ended” double-stranded DNA molecules lacking sticky ends. The resulting molecules are then treated with the restriction enzyme specific for the attached linker, generating cDNA molecules with sticky ends (step 8a). In a separate procedure, plasmid DNA is treated with the same restriction enzyme to produce the appropriate sticky ends (step 8b).
The plasmid vector and the collection of cDNAs, all containing complementary sticky ends, are then mixed and joined covalently by DNA ligase (Figure 6-17, step 9). The resulting DNA molecules are introduced into E. coli cells to generate individual clones; each clone carries a cDNA derived from a single mRNA.
Because different genes are transcribed at very different rates, cDNA clones corresponding to abundantly transcribed genes will be represented many times in a cDNA library, whereas cDNAs corresponding to infrequently transcribed genes will be extremely rare or not present at all. To have a reasonable chance of including clones corresponding to slowly transcribed genes, mammalian cDNA libraries must contain 106–107 individual recombinant clones.
