Recessive and Dominant Mutant Alleles Generally Have Opposite Effects on Gene Function
A fundamental genetic difference between experimental organisms is whether their cells carry two copies of each chromosome or only one copy of each chromosome. The former are referred to as diploid; the latter as haploid. Most complex multicellular organisms (e.g., fruit flies, mice, humans) are diploid, whereas some simple unicellular organisms are haploid. Some organisms, notably the yeast Saccharomyces cerevisiae, can exist in either haploid or diploid states. The normal cells of some organisms, both plants and animals, carry more than two copies of each chromosome and are thus designated polyploid. Moreover, cancer cells begin as diploid cells, but through the process of transformation into cancer cells, can gain extra copies of one or more chromosomes and are thus designated as aneuploid. However, our discussion of genetic techniques and analysis relates to diploid organisms, including diploid yeasts.
225
Although many different alleles of a gene might occur in different organisms in a population, any individual diploid organism will carry two copies of each gene and thus at most can have two different alleles. A diploid individual with two different alleles is heterozygous for a gene, whereas a diploid individual that carries two identical alleles is homozygous for a gene. A recessive mutant allele is defined as one in which both alleles must be mutant in order for the mutant phenotype to be observed; that is, the individual must be homozygous for the mutant allele to show the mutant phenotype. In contrast, the phenotypic consequences of a dominant mutant allele can be observed in a heterozygous individual carrying one mutant and one wild-
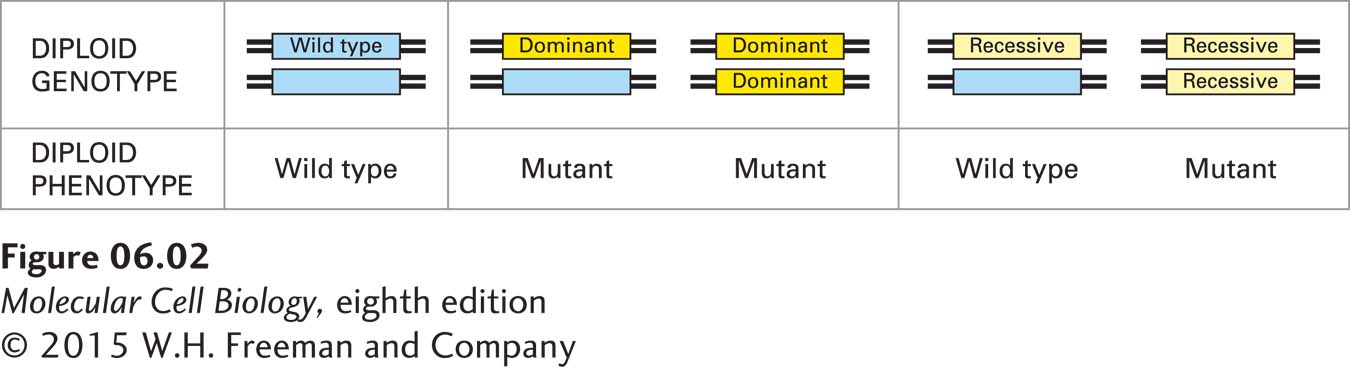
Whether a mutant allele is recessive or dominant provides valuable information about the function of the affected gene and the nature of the causative mutation. Recessive alleles usually result from a mutation that inactivates the affected gene, leading to a partial or complete loss of function. Such recessive mutations may remove part of the gene or the entire gene from the chromosome, disrupt expression of the gene, or alter the structure of the encoded protein, thereby altering its function. Conversely, dominant alleles are often the consequence of a mutation that causes some kind of gain of function. Such dominant mutations may increase the activity of the encoded protein, confer a new function on it, or lead to a new spatial or temporal pattern of expression.
In some rare cases, dominant mutations are associated with a loss of function. For instance, some genes are haplo-
Some alleles can exhibit both recessive and dominant properties. In such cases, statements about whether an allele is dominant or recessive must specify the phenotype. For example, the allele of the hemoglobin gene in humans designated Hbs has more than one phenotypic consequence. Individuals who are homozygous for this allele (Hbs/Hbs) have debilitating anemia caused by sickle-
A mutagen commonly used in experimental organisms is ethylmethane sulfonate (EMS). Although this mutagen can alter DNA sequences in several ways, its most common effect is to chemically modify guanine bases in DNA, ultimately leading to the conversion of a G·C base pair into an A·T base pair. Such an alteration in the sequence of a gene that involves only a single base pair is known as a point mutation. A silent point mutation causes no change in the amino acid sequence or activity of a gene’s encoded protein. However, observable phenotypic consequences due to changes in a protein’s activity can arise from point mutations that result in substitution of one amino acid for another (missense mutations) or introduction of a premature stop codon (nonsense mutations). Addition or deletion of bases may change the reading frame of a gene (frameshift mutations). Because alterations in the DNA sequence leading to a decrease in protein activity are much more likely than alterations leading to an increase or qualitative change in protein activity, mutagenesis usually produces many more recessive mutations than dominant mutations.