Genetic Suppression and Synthetic Lethality Can Reveal Interacting or Redundant Proteins
Two other types of genetic analysis can provide additional clues about how proteins that function in the same cellular process may interact with one another in the living cell. Both of these methods, which are applicable in many experimental organisms, involve the use of double mutants in which the phenotypic effects of one mutation are changed by the presence of a second mutation.
Suppressor Mutations The first type of analysis is based on genetic suppression. To understand this phenomenon, suppose that point mutations lead to structural changes in one protein (A) that disrupt its ability to associate with another protein (B) involved in the same cellular process. Similarly, mutations in protein B lead to small structural changes that inhibit its ability to interact with protein A. Assume, furthermore, that the normal functioning of proteins A and B depends on their interacting. In theory, a specific structural change in protein A might be suppressed by compensatory changes in protein B, allowing the mutant proteins to interact. In the rare cases in which such suppressor mutations occurred, strains carrying both mutant alleles would be normal, whereas strains carrying only one or the other mutant allele would have a mutant phenotype (Figure 6-9a).
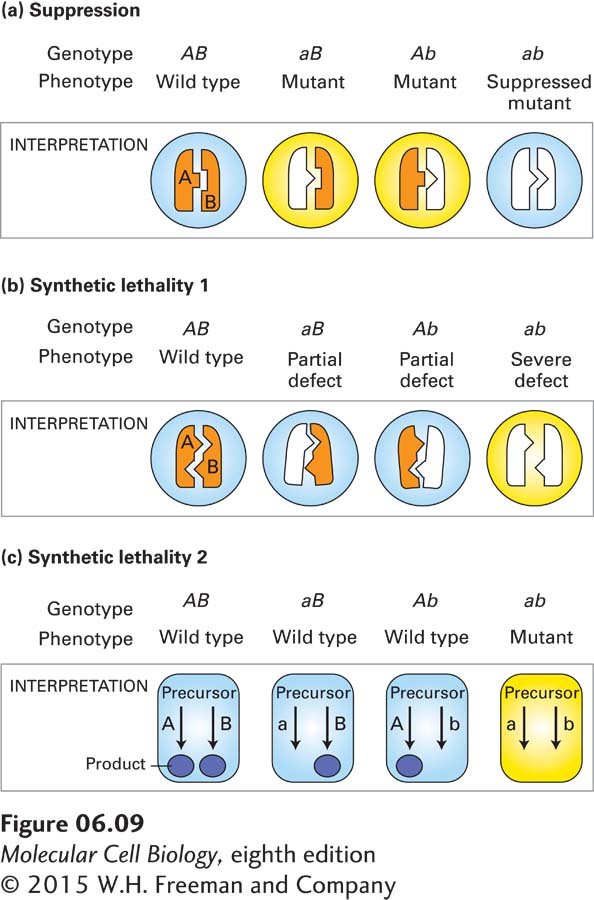
The observation of genetic suppression in yeast strains carrying a mutant actin allele (act1-
Synthetic Lethal Mutations Another phenomenon, called synthetic lethality, produces a phenotypic effect opposite to that of suppression. In this case, the deleterious effect of one mutation is greatly exacerbated (rather than suppressed) by a second mutation in a related gene. One situation in which such synthetic lethal mutations can occur is illustrated in Figure 6-9b. In this example, a heterodimeric protein is partially, but not completely, inactivated by mutations in either one of its nonidentical subunits. However, in double mutants carrying specific mutations in the genes encoding both subunits, little interaction between subunits occurs, resulting in severe phenotypic effects. Synthetic lethal mutations can also reveal nonessential genes whose encoded proteins function in redundant pathways for producing an essential cell component. As depicted in Figure 6-9c, if either pathway alone is inactivated by a mutation, the other pathway will be able to supply the needed product. However, if both pathways are inactivated at the same time, the essential product cannot be synthesized, and the double mutants will be nonviable.
232