Chromosome Painting and DNA Sequencing Reveal the Evolution of Chromosomes
Analysis of chromosomes from different species has provided considerable insight into how chromosomes evolved. For example, hybridization of chromosome paint probes for chromosome 16 of the tree shrew (Tupaia belangeri) to tree shrew metaphase chromosomes revealed the two copies of chromosome 16, as expected (Figure 8-39a). However, when the same chromosome paint probes were hybridized to human metaphase chromosomes, most of the probes hybridized to the long arm of chromosome 10 (Figure 8-39b). Further, when multiple probes for the long arm of human chromosome 10 with different fluorescent dye labels were hybridized to tree shrew metaphase chromosomes, these probes bound to sequences along tree shrew chromosome 16 in the same order in which they bind to human chromosome 10.
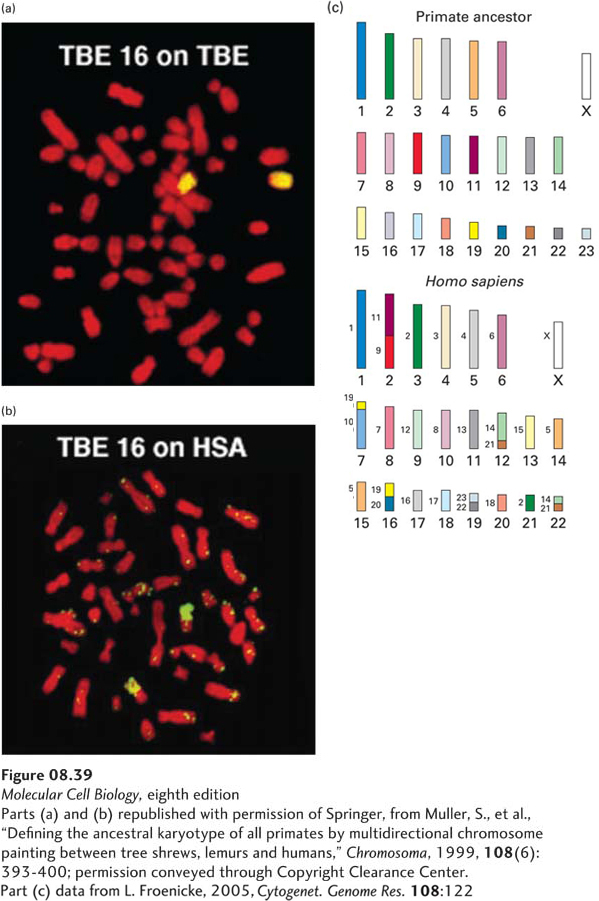
FIGURE 8-39 Evolution of primate chromosomes. (a) Chromosome paint probes (yellow) for chromosome 16 of the tree shrew (T. belangeri, distantly related to humans) hybridized to tree shrew metaphase chromosomes (red). (b) The same tree shrew chromosome 16 paint probes hybridized to human metaphase chromosomes. (c) Proposed evolution of human chromosomes (bottom) from the chromosomes of the common ancestor of all primates (top). The proposed common primate ancestor chromosomes are numbered according to their sizes, with each chromosome represented by a different color. The human chromosomes are also numbered according to their relative sizes and labeled with colors taken from the colors of the proposed common primate ancestor chromosomes from which they were derived. Small numbers to the left of the colored regions of the human chromosomes indicate the number of the ancestral chromosome from which the region was derived. Various human chromosomes were derived from the proposed chromosomes of the common primate ancestor without significant rearrangements (e.g., human chromosome 1); by fusion (e.g., human chromosome 2 by fusion of ancestral chromosomes 9 and 11); by breakage (e.g., human chromosomes 14 and 15 by breakage of ancestral chromosome 5); or by chromosomal translocations (e.g., a reciprocal translocation between ancestral chromosomes 14 and 21 generated human chromosomes 12 and 22).
[Parts (a) and (b) republished with permission of Springer, from Muller, S., et al., “Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans,” Chromosoma, 1999, 108(6):393-400; permission conveyed through Copyright Clearance Center. Part (c) data from L. Froenicke, 2005, Cytogenet. Genome Res. 108:122.]
These results indicate that during the evolution of humans and tree shrews from a common ancestor that lived as recently as 85 million years ago, a long, continuous DNA sequence on one of the ancestral chromosomes became chromosome 16 in tree shrews, but evolved into the long arm of chromosome 10 in humans. The phenomenon of genes occurring in the same order on a chromosome in two different species is referred to as conserved synteny (derived from Latin for “on the same ribbon”). The presence of two or more genes in a common chromosomal region in two or more species indicates a conserved syntenic segment.
The relationships between the chromosomes of many primates have been determined by cross-species application of chromosome paint probes, as shown for human and tree shrew in Figure 8-39a, b. Using these relationships, as well as higher-resolution analyses of regions of synteny by DNA sequencing and other methods, it has been possible to propose the karyotype of the common ancestor of all primates based on the minimum number of chromosomal rearrangements necessary to generate the regions of synteny in chromosomes of contemporary primates.
Human chromosomes are thought to have been derived from a common primate ancestor with 23 autosomes plus the X and Y sex chromosomes by several different mechanisms (Figure 8-39c). Some human chromosomes were derived without large-scale rearrangements of chromosome structure. Others are thought to have evolved by breakage of an ancestral chromosome into two chromosomes or, conversely, by fusion of two ancestral chromosomes. Still other human chromosomes appear to have been generated by exchanges of parts of the arms of distinct chromosomes; that is, by reciprocal translocation involving two ancestral chromosomes. Analysis of regions of conserved synteny between the chromosomes of many mammals indicates that chromosomal rearrangements by breakage, fusion, and translocations occurred rarely in mammalian evolution, about once every 5 million years. When such chromosomal rearrangements did occur, they very likely contributed to the evolution of new species that could not interbreed with the species from which they evolved.
Chromosomal rearrangements similar to those inferred for the primate lineage have been inferred for other groups of related organisms, including the invertebrate, plant, and fungus lineages. The excellent agreement between predictions of evolutionary relationships based on analysis of syntenic regions of chromosomes from organisms with related morphology (i.e., among mammals, among insects with similar body organization, among similar plants, etc.) and evolutionary relationships based on the fossil record and on the extent of divergence of DNA sequences for homologous genes is a strong argument for the validity of evolution as the process that generated the diversity of contemporary organisms.
