General Transcription Factors Position RNA Polymerase II at Start Sites and Assist in Initiation
Initiation of transcription by RNA polymerase II requires several initiation factors. These initiation factors position Pol II molecules at transcription start sites and help to separate the DNA strands so that the template strand can enter the active site of the enzyme. They are called general transcription factors because they are required at most, if not all, promoters of genes transcribed by RNA polymerase II. These proteins are designated TFIIA, TFIIB, and so on, and most are multimeric proteins. The largest is TFIID, which consists of a single 38-kDa TATA box–binding protein (TBP) and 13 TBP-associated factors (TAFs). General transcription factors with similar activities and homologous sequences are found in all eukaryotes. The complex of Pol II and its general transcription factors bound to a promoter and ready to initiate transcription is called a preinitiation complex (PIC). Figure 9-19 summarizes the current model for the stepwise assembly of the Pol II transcription preinitiation complex on a promoter containing a TATA box.
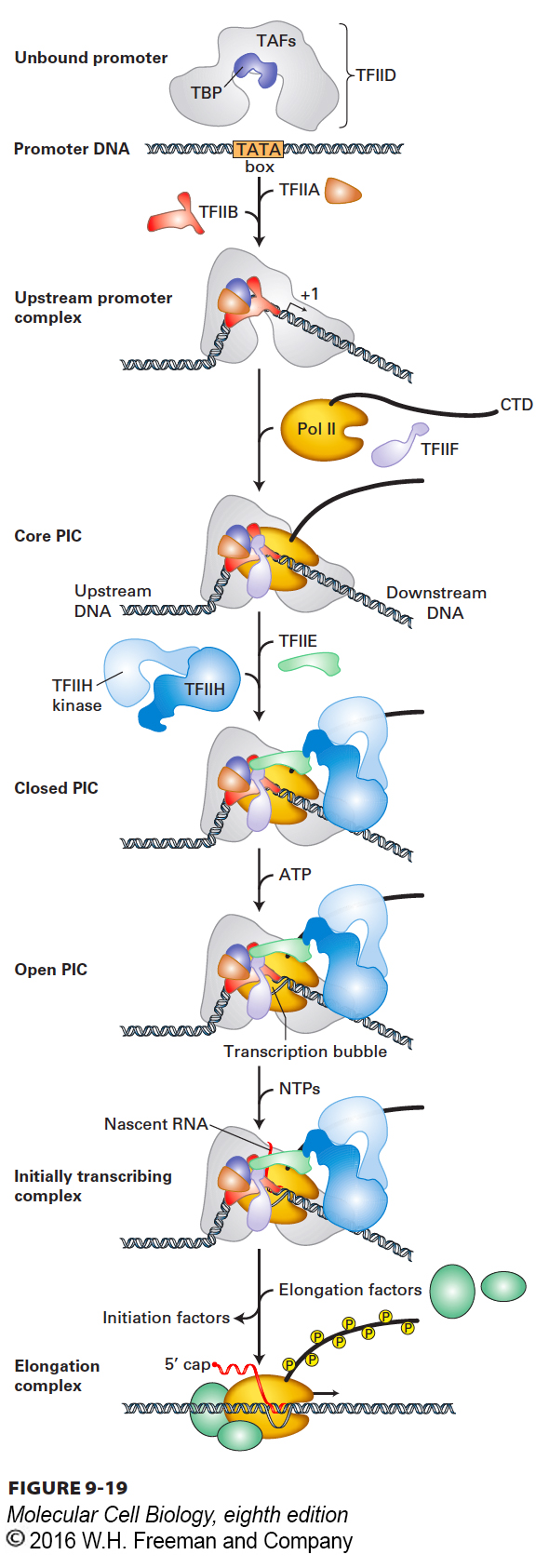
FIGURE 9-19 Model for the sequential assembly of an RNA polymerase II preinitiation complex. The indicated general transcription factors and purified RNA polymerase II (Pol II) bind sequentially to TATA box DNA to form a preinitiation complex (PIC). ATP hydrolysis then provides the energy for the unwinding of DNA at the transcription start site by a TFIIH helicase subunit that pushes downstream DNA into the polymerase. The DNA is held in position in the PIC by binding of the TATA box by the TBP subunit of TFIID, and the resulting strain on the structure of the duplex DNA assists the N-terminal region of TFIIB and Pol II to melt the DNA at the transcription start site, forming the transcription bubble. As Pol II initiates transcription in the resulting open complex, the polymerase transcribes away from the promoter, its CTD becomes phosphorylated by the TFIIH kinase domain, and the general transcription factors dissociate from the promoter. See S. Sainsbury, C. Berrnecky, and P. Cramer, 2015, Nat. Rev. Mol. Cell Biol. 16:129.
The TBP subunit of TFIID is the first protein to bind to a TATA box promoter. All eukaryotic TBPs analyzed to date have very similar C-terminal domains of 180 residues. This domain of TBP folds into a saddle-shaped structure; the two halves of the molecule exhibit an overall dyad symmetry but are not identical. TBP interacts with the minor groove in DNA, bending the helix considerably (see Figure 5-5). The DNA-binding surface of TBP is conserved in all eukaryotes, explaining the high conservation of the TATA box promoter element (see Figure 9-16).
Once TFIID has bound to the TATA box, TFIIA and TFIIB can bind. TFIIA is a heterodimer larger than TBP, and TFIIB is a monomeric protein, slightly smaller than TBP. TFIIA associates with TBP and DNA on the upstream side of the TBP–TATA box complex. The C-terminal domain of TFIIB makes contact with both TBP and DNA on either side of the TATA box. During transcription initiation, its N-terminal domain is inserted into the RNA exit channel of RNA polymerase II (see Figure 9-12c). The TFIIB N-terminal domain assists Pol II in melting the DNA strands at the transcription start site and interacts with the template strand near the Pol II active site. Following TFIIB binding, a preformed complex of TFIIF (a heterodimer of two different subunits in mammals) and Pol II binds, positioning the polymerase over the start site. Two more general transcription factors must bind before the DNA duplex can be separated to expose the template strand. First to bind is TFIIE, a heterodimer of two different subunits. TFIIE creates a docking site for TFIIH, another multimeric factor containing 10 different subunits. Binding of TFIIH completes assembly of the transcription preinitiation complex (see Figure 9-19).
Figure 9-20 shows a cryoelectron microscopic image of a yeast (S. cerevisiae) preinitiation complex assembled in vitro from purified RNA polymerase II and general transcription factors with TBP in place of the complete TFIID complex—a total of thirty-three polypeptides with a mass of 1.5 megadaltons (MDa)—about the size of a ribosomal subunit. Such elaborate preinitiation complexes assemble at the promoters of every protein-coding gene expressed by a eukaryotic cell.
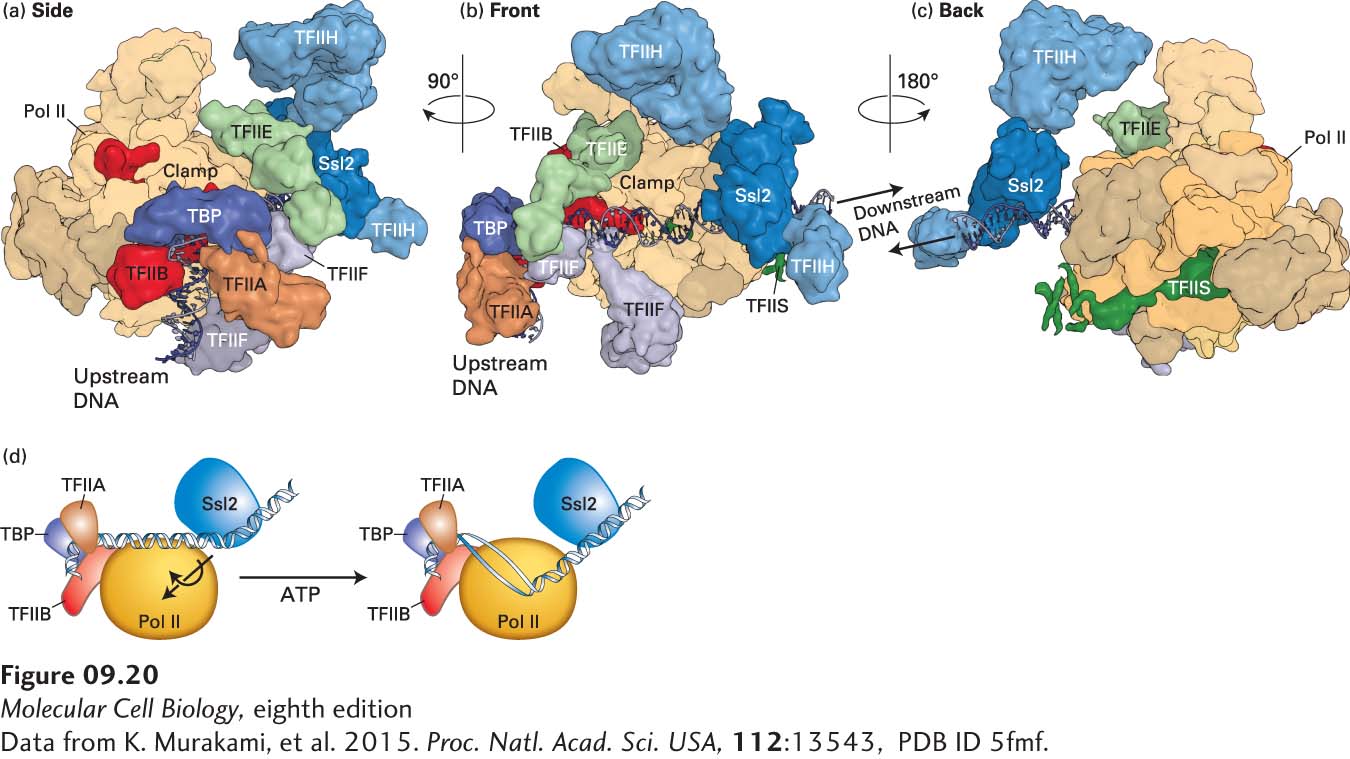
FIGURE 9-20 Model Of The Yeast preinitiation complex based on cryoelectron microscopy and fitting of known protein x-ray crystal structures. (a-c) Three views of the nearly complete PIC. The relative positions of Pol II and most of the GTFs are observed, but only about 50% of the mass of TFIIH is depicted because a large part of the mass of TFIIH is highly flexible and consequently could not be accurately determined by cryo-EM. Also high resolution structures have not been determined for many of the TFIIH subunits, and consequently could not be fitted to the TFIIH mass detected by cryo-EM. However, the interaction between DNA at the downstream side of the Pol II cleft and the TFIIH Ssl2 helicase subunit required to melt promoter DNA is clearly visualized in (b) and (c). In (c), the interaction between TFIIH and TFIIE is not visualized because of the low resolution of the complex in this region. TFIIS is a Pol II elongation factor added to stabilize the PIC. (d) Model of entry of the template strand into the floor of the cleft where RNA polymerization is catalyzed. The Ssl2 helicase pushes DNA that is bound upstream to TBP, TFIIB, and TFIIA, creating torsional stress that contributes to transcription bubble melting.
[Data from K. Murakami, et al. 2015. Proc. Natl. Acad. Sci. USA, 112:13543, PDB ID 5fmf.]
The helicase activity of one of the core TFIIH subunits (Ssl2 in yeast; see Figure 9-20d) uses energy from ATP hydrolysis to help unwind the DNA duplex at the start site, allowing Pol II to form an open complex in which the DNA duplex surrounding the start site is melted and the template strand is bound at the polymerase active site. As the polymerase transcribes away from the promoter region, the N-terminal domain of TFIIB is released from the RNA exit channel as the 5′ end of the nascent RNA enters it. Three TFIIH subunits form a kinase module (TFIIH kinase in Figure 9-19) that phosphorylates the Pol II CTD multiple times on serine 5 (underlined) of the Tyr-Ser-Pro-Thr-Ser-Pro-Ser repeat that constitutes the CTD. As we will discuss further in Chapter 10, a multiply phosphorylated CTD is a docking site for the enzymes that form the cap structure (see Figure 5-14) on the 5′ end of an RNA transcribed by RNA polymerase II. In the minimal in vitro transcription assay with TBP substituted for the full TFIID complex and purified RNA polymerase II, TBP remains bound to the TATA box as the polymerase transcribes away from the promoter region, but the other general transcription factors dissociate.
Remarkably, the first subunits of TFIIH to be cloned from humans were identified because mutations in them cause defects in the repair of damaged DNA, such as a base with a covalently linked mutagen or a UV-induced thymine-thymine dimer (see Figure 5-37). In normal individuals, when a transcribing RNA polymerase becomes stalled at a region of damaged template DNA, the core TFIIH complex, lacking the three subunits of the kinase domain (see Figure 9-19) but including the helicase subunit mentioned above, recognizes the stalled polymerase and then associates with other proteins that function with TFIIH in repairing the damaged DNA region. In patients with mutant forms of these TFIIH subunits, such repair of damaged DNA in transcriptionally active genes is impaired. As a result, affected individuals have extreme skin sensitivity to sunlight (a common cause of DNA damage through the generation of thymine-thymine dimers) and exhibit a high incidence of cancer. Consequently, these subunits of TFIIH serve two functions in the cell, one in the process of transcription initiation and a second in the repair of DNA. Depending on the severity of the defect in TFIIH function, these individuals may suffer from diseases such as xeroderma pigmentosum (see Chapter 24) and Cockayne syndrome (see Chapter 5).
The TAF subunits of TFIID function in initiating transcription from promoters that lack a TATA box. For instance, some TAF subunits contact the initiator element in promoters in which it occurs; their function probably explains how such sequences can replace a TATA box (see Figure 9-16). Additional TFIID TAF subunits can bind to a consensus sequence, A/G-G-A/T-C/T-G/A/C, that is centered about 30 bp downstream from the transcription start site in many genes that lack a TATA box promoter. Because of its position, this regulatory sequence is called the downstream promoter element (DPE) (see Figure 9-16). The DPE facilitates transcription of TATA-less genes that contain it by increasing TFIID binding. In addition, an α helix of TFIIB binds to the major groove of DNA upstream of the TATA box, and the strongest promoters contain the optimal sequence for this interaction, called the TFIIB recognition element (BRE) (see Figure 9-16).
Chromatin immunoprecipitation assays (see Figure 9-18) using antibodies to TBP show that it binds in the region between the divergent transcription start sites in CpG island promoters. Consequently, the same general transcription factors are probably required for initiation from the weaker CpG island promoters as for initiation from promoters containing a TATA box. The absence of the promoter elements summarized in Figure 9-16 may account for the divergent transcription from multiple transcription start sites observed from CpG island promoters, since cues from the DNA sequence are not present to correctly orient the preinitiation complex. TFIID and the other general transcription factors may choose among alternative, nearly equivalent weak binding sites in CpG island promoters, which may explain the low frequency of transcription initiation as well as the alternative transcription start sites in divergent directions generally observed from this class of promoters.

