Most Eukaryotic Genes Are Regulated by Multiple Transcription-Control Elements
Initially, enhancers and promoter-proximal elements were thought to be distinct types of transcription-control elements. However, as more enhancers and promoter-proximal elements were analyzed, the distinctions between them became less clear. For example, both types of elements can generally stimulate transcription even when inverted, and both types are often cell-type-specific. The general consensus now is that a spectrum of control elements regulates transcription by RNA polymerase II. At one extreme are enhancers, which can stimulate transcription from a promoter tens of thousands of base pairs away. At the other extreme are promoter-proximal elements, such as the upstream elements controlling the HSV-I tk gene, which lose their influence when moved 30–50 bp farther from the promoter. Researchers have identified a large number of transcription-control elements that can stimulate transcription from distances between these two extremes.
Figure 9-23a summarizes the locations of transcription-control sequences for a hypothetical mammalian gene with a promoter containing a TATA box. The transcription start site encodes the first (5′) nucleotide of the first exon of an mRNA, the nucleotide that is capped. In addition to the TATA box at about −31 to −26, promoter-proximal elements, which are relatively short (~6–10 bp), are located within the first 200 bp either upstream or downstream of the start site. Enhancers, in contrast, are usually about 50–200 bp long and are composed of multiple elements of about 6–10 bp. Enhancers may be located up to 50 kb or more upstream or downstream from the start site or within an intron. Like the Pax6 gene, many mammalian genes are controlled by multiple enhancer regions that function in different types of cells.
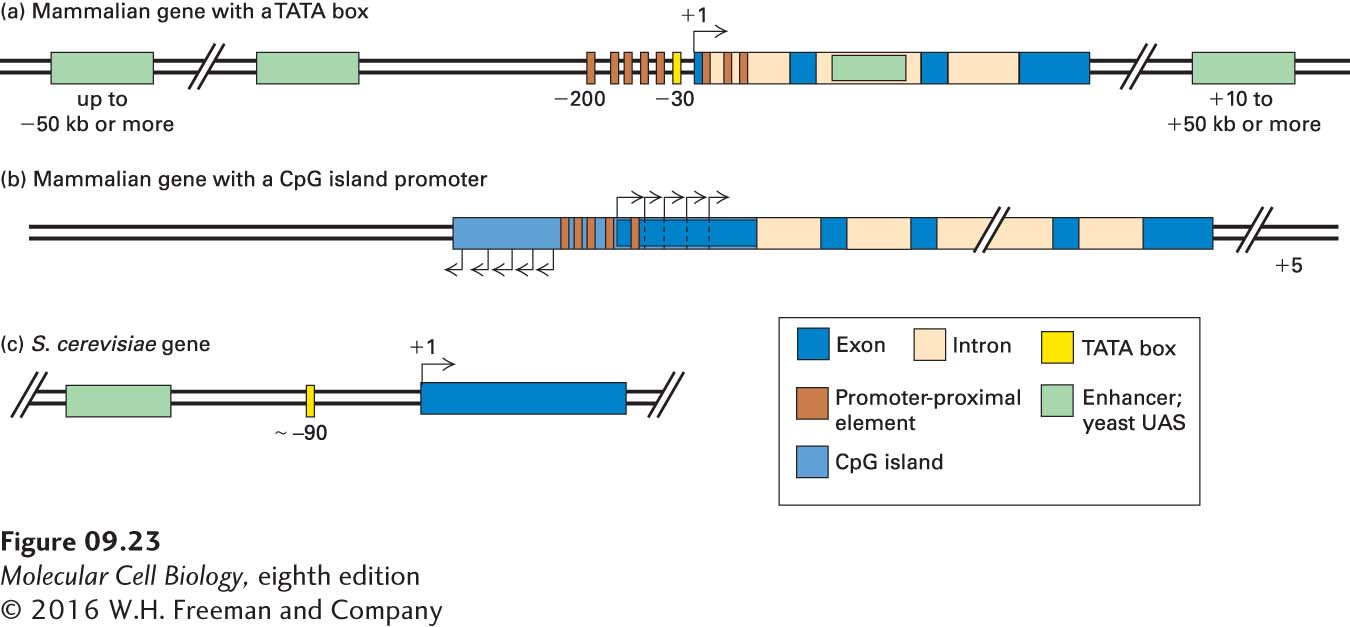
FIGURE 9-23 General organization of control elements that regulate gene expression in multicellular eukaryotes and yeast. (a) Mammalian genes with a TATA box promoter are regulated by promoter-proximal elements and enhancers. The promoter elements shown in Figure 9-16 position RNA polymerase II to initiate transcription at the start site and influence the rate of transcription. Enhancers may be either upstream or downstream and as far away as hundreds of kilobases from the transcription start site. In some cases, enhancers lie within introns. Promoter-proximal elements are found upstream and downstream of transcription start sites at equal frequency in mammalian genes. (b) For mammalian genes with a CpG island promoter, transcription initiates at several sites in both the sense and antisense directions from the ends of the CpG-rich region. Transcripts in the sense direction are elongated and are processed into mRNAs by RNA splicing. These genes express mRNAs with alternative 5′ exons determined by the transcription start site. Genes with CpG island promoters contain promoter-proximal control elements. Currently, it is not clear whether they are also regulated by distant enhancers. (c) Most S. cerevisiae genes contain only one regulatory region, called an upstream activating sequence (UAS), and a TATA box, which is about 90 bp upstream from the transcription start site.
Figure 9-23b summarizes the promoter region of a mammalian gene with a CpG island promoter. About 70 percent of mammalian genes are expressed from CpG island promoters, usually at much lower levels than genes with TATA box promoters. Multiple alternative transcription start sites are used, generating mRNAs with alternative 5′ ends for the first exon derived from each start site. Transcription occurs in both directions, but Pol II molecules transcribing in the sense direction are elongated to 1 kb or more, much more efficiently than transcripts in the antisense direction.
In the important model organism Saccharomyces cerevisiae (budding yeast), genes are closely spaced (see Figure 8-4b), and few genes contain introns. In this organism, enhancers, which are referred to as upstream activating sequences (UASs), usually lie within 200 bp upstream of the promoters of the genes they regulate. Most yeast genes contain only one UAS. In addition, S. cerevisiae genes contain a TATA box about 90 bp upstream from the transcription start site (Figure 9-23c).
