Chromatin-Remodeling Complexes Help Activate or Repress Transcription
In addition to histone acetylase complexes, multiprotein chromatin-remodeling complexes are required for activation at many promoters. The first of these complexes characterized was the yeast SWI/SNF chromatin-remodeling complex. One of the SWI/SNF subunits has homology to DNA helicases, enzymes that use energy from ATP hydrolysis to disrupt interactions between base-paired nucleic acids or between nucleic acids and proteins. In vitro, the SWI/SNF complex is thought to pump or push DNA into the nucleosome so that DNA bound to the surface of the histone octamer transiently dissociates from the surface and translocates, causing the nucleosomes to “slide” along the DNA. The net result of such chromatin remodeling is to facilitate the binding of transcription factors to specific DNA sequences in chromatin. Many activation domains bind to such chromatin-remodeling complexes, and this binding stimulates in vitro transcription from chromatin templates in which the DNA is associated with histone octamers. Thus the SWI/SNF complex represents another type of co-activator complex. The experiment shown in Figure 9-38 demonstrates dramatically how an activation domain can cause decondensation of a region of chromatin. This decondensation results from association of the activation domain with chromatin-remodeling and histone acetylase complexes.
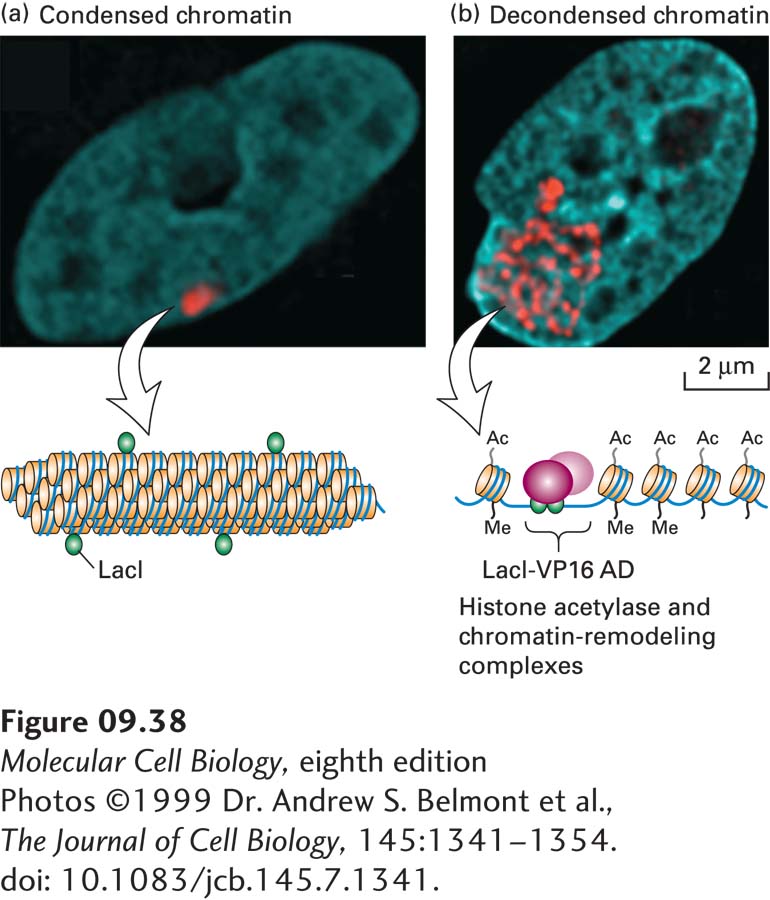
FIGURE 9-38 Expression of fusion proteins demonstrates chromatin decondensation in response to an activation domain. A cultured hamster cell line was engineered to contain multiple copies of a tandem array of E. coli lac operator sequences integrated into a chromosome in a region of heterochromatin. (a) When an expression vector for the lac repressor (LacI) was transfected into these cells, lac repressor bound to the lac operator sites could be visualized in a region of condensed chromatin using an antibody against the lac repressor (red). DNA was visualized by staining with DAPI (blue), revealing the nucleus. A diagram of condensed chromatin is shown below. (b) When LacI fused to an activation domain was transfected into these cells, staining as in (a) revealed that the activation domain causes this region of chromatin to decondense into a thinner chromatin fiber that fills a much larger volume of the nucleus. A diagram of a region of decondensed chromatin with bound LacI fusions to the VP16 activation domain (AD) and associated chromatin remodeling and histone acetylase complexes is shown below.
[Photos ©1999 Dr. Andrew S. Belmont et al., The Journal of Cell Biology, 145:1341–1354. doi: 10.1083/jcb.145.7.1341.]
Chromatin-remodeling complexes are required for many processes involving DNA in eukaryotic cells, including transcriptional control, DNA replication, recombination, and DNA repair. Several types of chromatin-remodeling complexes are found in eukaryotic cells, all with homologous DNA helicase domains. SWI/SNF complexes and related chromatin-remodeling complexes in multicellular organisms contain subunits with bromodomains that bind to acetylated histone tails. Consequently, SWI/SNF complexes remain associated with activated, acetylated regions of chromatin, presumably maintaining them in a decondensed conformation. Chromatin-remodeling complexes can also participate in transcriptional repression. These complexes bind to the repression domains of repressors and contribute to repression, presumably by folding chromatin into condensed structures. Much remains to be learned about how this important class of proteins alters chromatin structure to influence gene expression and other processes.

