Chapter 13. Investigating Translocation of Prolactin
Introduction

Analyze the Data 13-1: Investigating Translocation of Prolactin
Imagine that you are evaluating the early steps in the translocation and processing of the secretory protein prolactin. By using an experimental approach similar to that shown in Figure 13-7, you can use truncated prolactin mRNAs to control the length of nascent prolactin polypeptides that are synthesized. When prolactin mRNA that lacks a stop codon is translated in vitro, the newly synthesized polypeptide, ending with the last codon included on the mRNA, will remain attached to the ribosome, thus allowing a polypeptide of defined length to extend from the ribosome. You have generated a set of mRNAs that encode segments of the N-terminus of prolactin of increasing length, and each mRNA can be translated in vitro by a cytosolic translation extract containing ribosomes, tRNAs, aminoacyl-tRNA synthetases, GTP, and translation initiation and elongation factors. When radiolabeled amino acids are included in the translation mixture, only the polypeptide encoded by the added mRNA will be labeled. After completion of translation, each reaction mixture is resolved by SDS-polyacrylamide gel electrophoresis, and the labeled polypeptides are identified by autoradiography.
Question
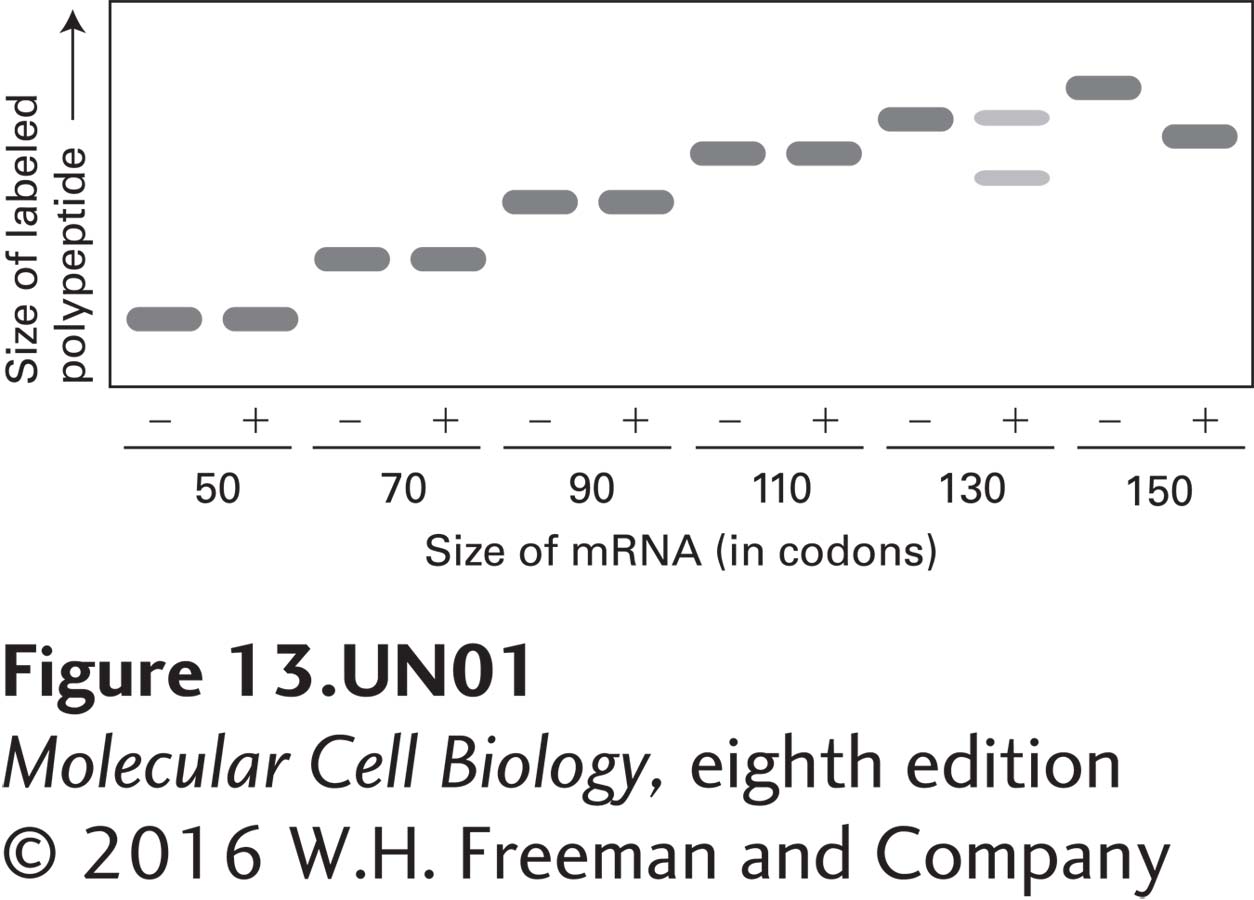
a. The autoradiogram depicted below shows the results of an experiment in which each translation reaction was carried out either in the presence (+) or the absence (−) of microsomal membranes. Based on the gel mobility of peptides synthesized in the presence or absence of microsomes, deduce how long the prolactin nascent chain must be in order for the prolactin signal peptide to enter the ER lumen and to be cleaved by signal peptidase. (Note that microsomes carry significant quantities of signal recognition particles weakly bound to the membranes.)
Question
b. Given this length, what can you conclude about the conformational state(s) of the nascent prolactin polypeptide when it is cleaved by signal peptidase? The following lengths will be useful for your calculation: the prolactin signal sequence is cleaved after amino acid 31; the channel within the ribosome occupied by a nascent polypeptide is about 150 Å long; a membrane bilayer is about 50 Å thick; in polypeptides with an α-helical conformation, one residue extends 1.5 Å, whereas in fully extended polypeptides, one residue extends about 3.5 Å.
5c/DQOGODaA5ROLTyR8YjhQwCA5aDGWkfQpA+l2yJO2PaWJUX7+Kfg==Question
4ttWuwbM4MLs5kBNdPk9urp1whz86lXNB340563ZiKraJqT9EanyeSrN14KgGMqrvWetbtBPcqYTwko0HCv2VKEh3PjL+mUBad0X0jeyNijyyGWMLGc9DgDoFpM4mQviuUcW1iDa+MWTejf28Vo2Cu1cnddOc71OMwx67b3dlIxtHnRTlnG5XDyN2JxoP8xPClDAQYt150pH5jNVLB1Kme2Xqlmhfqmg/odGUCYExeMfhJ2rHrdUumDFZkYnePeyb71W2JvLgEb/JU4Z//v4E+Vp7PgvygQDGOOVKjN0X9BiCFyAJis5ntIiJkeoeSqO4MBcfS0PCyU4A1xE4lW8NwXohtUUH4fTfFj1rl84crXBJczAvohyps9PJxLi3FrMahY+NwNcJJyLncbfJqP+DIRM69rIpLmn5HSdNFP5mE8lm+ZGbuPtuuQFZOdsf42ke3CJG7Z7N/UFDHwdIiNhKy6IZPJr5HRHd3fGbO+apZHPtDmj7uwAZbM1ZT8IxeF5+pC6s8RSZdax6a/o6BHlr9StZIqhzF+q51UARrMvo0cLZfxzQuRWw0zTFWuHTREnir3VqPKoldZv7ZG/M01EqyuGrJQ=Question
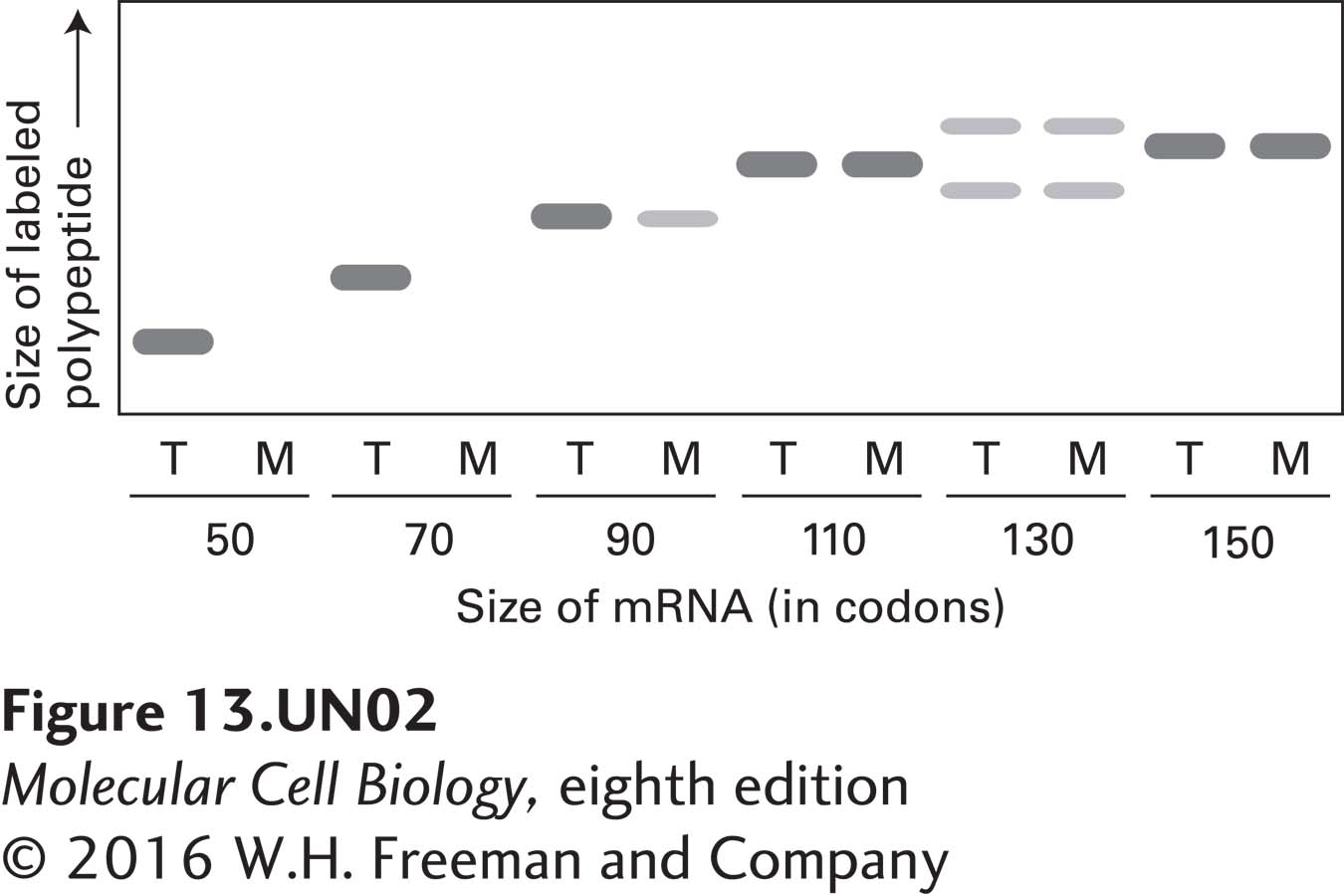
d. In another experiment, each translation reaction was carried out in the presence of microsomes, and the microsomal membranes and bound ribosomes were then separated from free ribosomes and soluble proteins by centrifugation. For each translation reaction, both the total reaction (T) and the membrane fraction (M) were resolved in neighboring gel lanes. Based on the amounts of labeled polypeptide in the membrane fractions in the autoradiogram depicted below, deduce how long the prolactin nascent chain must be in order for ribosomes engaged in translation to engage the SRP and thereby become bound to microsomal membranes.
Activity results are being submitted...