HOW DO WE KNOW?
How many kinds of bacterium live in the oceans?
BACKGROUND The oceans are full of bacteria—by some estimates, the sea harbors more than 1029 bacterial cells. How diverse are these bacterial communities?
METHOD 1 Different types of bacterium can be recognized by the unique nucleotide sequences of their genes. In fact, individual types of bacterium can be identified on the basis of nucleotide sequence in a relatively small region of the gene for the small subunit of ribosomal RNA (rRNA). This molecular “barcode” can be sequenced quickly and accurately for samples of DNA drawn from the environment.
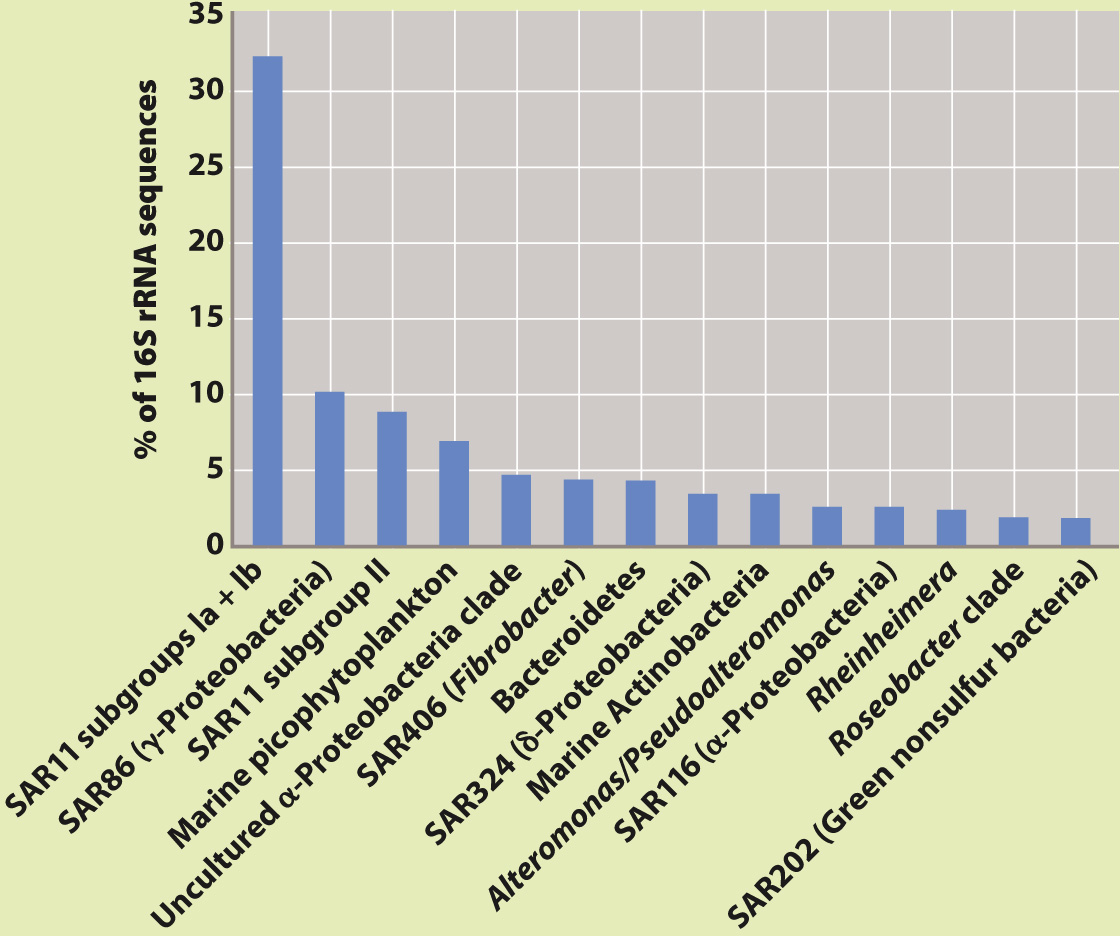
FIG. 26.13 Relative abundance of different bacterial groups in samples from the Sargasso Sea, based on percent representation of small subunit rRNA sequences. Note the abundance of SAR11, only recently characterized.
Using this strategy, scientists associated with the International Census of Marine Life obtained samples of seawater and seafloor sediment from deep-ocean environments. In the laboratory, the rRNA sequences were amplified by PCR, separated by gel electrophoresis, and sequenced, thus providing a library of tags that document bacterial diversity.
METHOD 2 In another set of experiments, scientists collected samples of seawater from the Sargasso Sea, and from these they amplified whole genomes—more than a billion nucleotides’ worth. They used small-subunit rRNA and other genes to characterize species richness, and also characterized a large assortment of additional sequence data to understand genetic and physiological diversity.
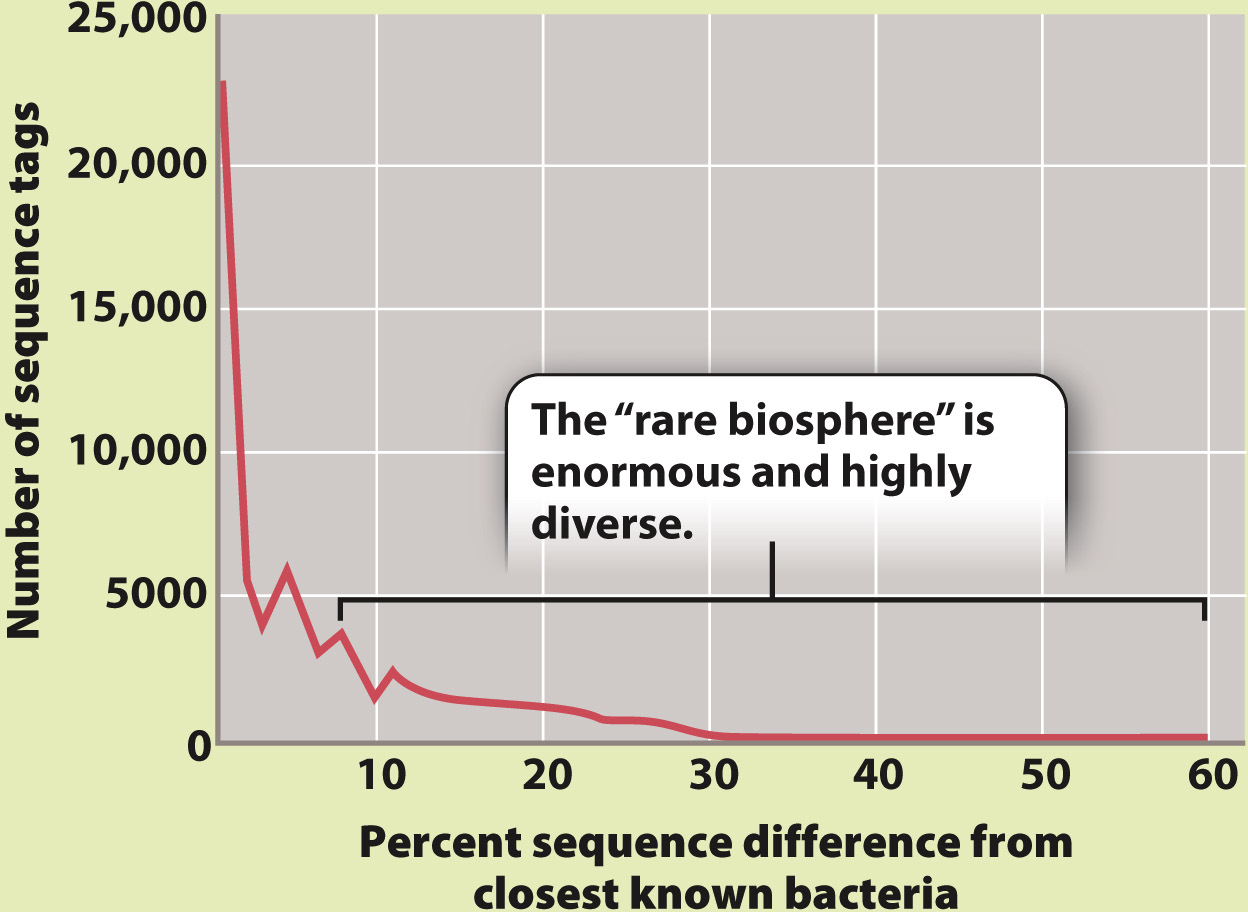
Computer-aided comparison of sequences from the world’s oceans shows that about 75% of all sequences collected are similar to other sequences in the library of sequence tags. However, 25% are distinctive, forming a rare but enormously diverse pool of bacterial types.
RESULTS Both surveys found that the bacterial diversity of marine environments is much higher than had been thought. The barcode survey (method 1) found as many as 20,000 distinct types of bacteria in a single liter of seawater and estimated that as many as 5–10 million different microbes may live in the world’s oceans. Some of these bacteria are abundant and widespread, but most are rare. The genomic survey (method 2) of the Sargasso Sea found more than 1800 distinct bacteria in sea-surface samples and, remarkably, identified more than 1.2 million previously unknown genes within these genomes.
CONCLUSION The bacterial diversity of the oceans is huge and still largely unexplored. Both barcode and genomic surveys are continuing, and promise a more nearly complete accounting of marine diversity. Together, these surveys will help us understand how marine bacteria function and how they sort into communities.
SOURCES Venter, J. C., et al. 2004. “Environmental Genome Shotgun Sequencing of the Sargasso Sea.” Science 304:66–74; Sogin, M. L., et al. 2006. “Microbial Diversity in the Deep Sea and the Underexplored ‘Rare Biosphere’” Proceedings of the National Academy of Sciences USA 103:12115–12120.

