The polymerase chain reaction selectively amplifies regions of DNA.
DNA that exists in the nuclei of your cells is present in two copies per cell, except for DNA in the X and Y chromosomes in males, which are each present in only one copy per cell in males. In the laboratory, it is very difficult to manipulate or visualize a sample containing just one or two copies of a DNA molecule. Instead, researchers typically work with many identical copies of the DNA molecule they are interested in. A common method for making copies of a piece of DNA is the polymerase chain reaction (PCR), which allows a targeted region of a DNA molecule to be replicated (or amplified) into as many copies as desired. PCR is both selective and highly sensitive, so it is used to amplify and detect small quantities of nucleic acids, such as HIV in blood-
258
259
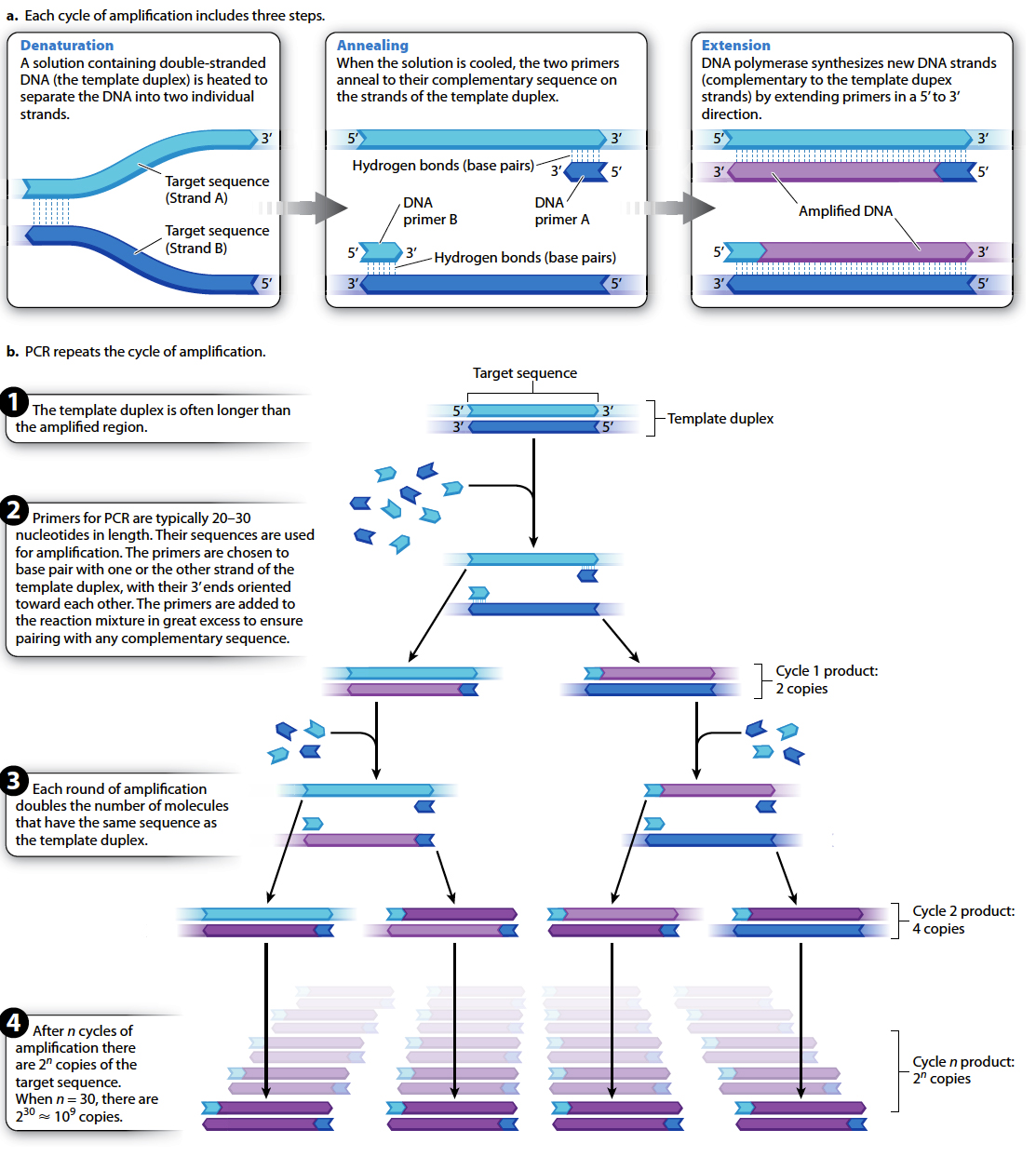
The principles of PCR are illustrated in Fig. 12.13. Because the PCR reaction is essentially a DNA synthesis reaction, it requires the same basic components used by the cell to replicate its DNA. In this case, the procedure takes place in a small plastic tube containing a solution that includes four essential components:
Template DNA. At least one molecule of double-
stranded DNA containing the region to be amplified serves as the template for amplification. DNA polymerase. The enzyme DNA polymerase is used to replicate the DNA.
All four deoxynucleoside triphosphates. Deoxynucleoside triphosphates with the bases A, T, G, or C are needed as building blocks for the synthesis of new DNA strands.
Two primers. Two short sequences of single-
stranded DNA are required for the DNA polymerase to start synthesis. Enough primer is added so that the number of primer DNA molecules is much greater than the number of template DNA molecules.
The primer sequences are oligonucleotides (oligos is the Greek word for “few”) produced by chemical synthesis and are typically 20–
PCR creates new DNA fragments in a cycle of three steps, as shown in Fig. 12.13a. The first step, denaturation, involves heating the solution in a plastic tube to a temperature just short of boiling so that the individual DNA strands of the template separate (or “denature”) as a result of the breaking of hydrogen bonds between the complementary bases. The second step, annealing, begins as the solution is cooled. Because of the great excess of primer molecules, the two primers bind (or “anneal”) to their complementary sequence on the DNA (rather than two strands of the template duplex coming back together). In the final step, extension, the solution is heated to the optimal temperature for DNA polymerase and the polymerase elongates (or “extends”) each primer with the deoxynucleoside triphosphates.
After sufficient time to allow new DNA synthesis, the solution is heated again, and the cycle of denaturation, annealing, and extension is repeated over and over, as indicated in Fig. 12.13b, usually for 25–
Although PCR is elegant in its simplicity, the DNA polymerase enzymes from many species (including humans) irreversibly lose both structure and function at the high temperature required to separate the DNA strands. At each cycle, you would have to open the tube and add fresh DNA polymerase. This is possible, and in fact it was how PCR was done when the technique was first developed, but the procedure is time consuming and tedious. To solve this problem, we now use DNA polymerase enzymes that are heat-