Electrophoresis separates DNA fragments by size.
PCR amplification does not always work as you might expect. Sometimes the primers have the wrong sequence and so fail to anneal properly; sometimes they anneal to multiple sites and several different fragments are amplified. To determine whether or not PCR has yielded the expected product, a researcher must determine the size of the amplified DNA molecules. Usually, the researcher knows what the size of the correctly amplified fragment should be, making it possible to compare the expected size to the actual size.
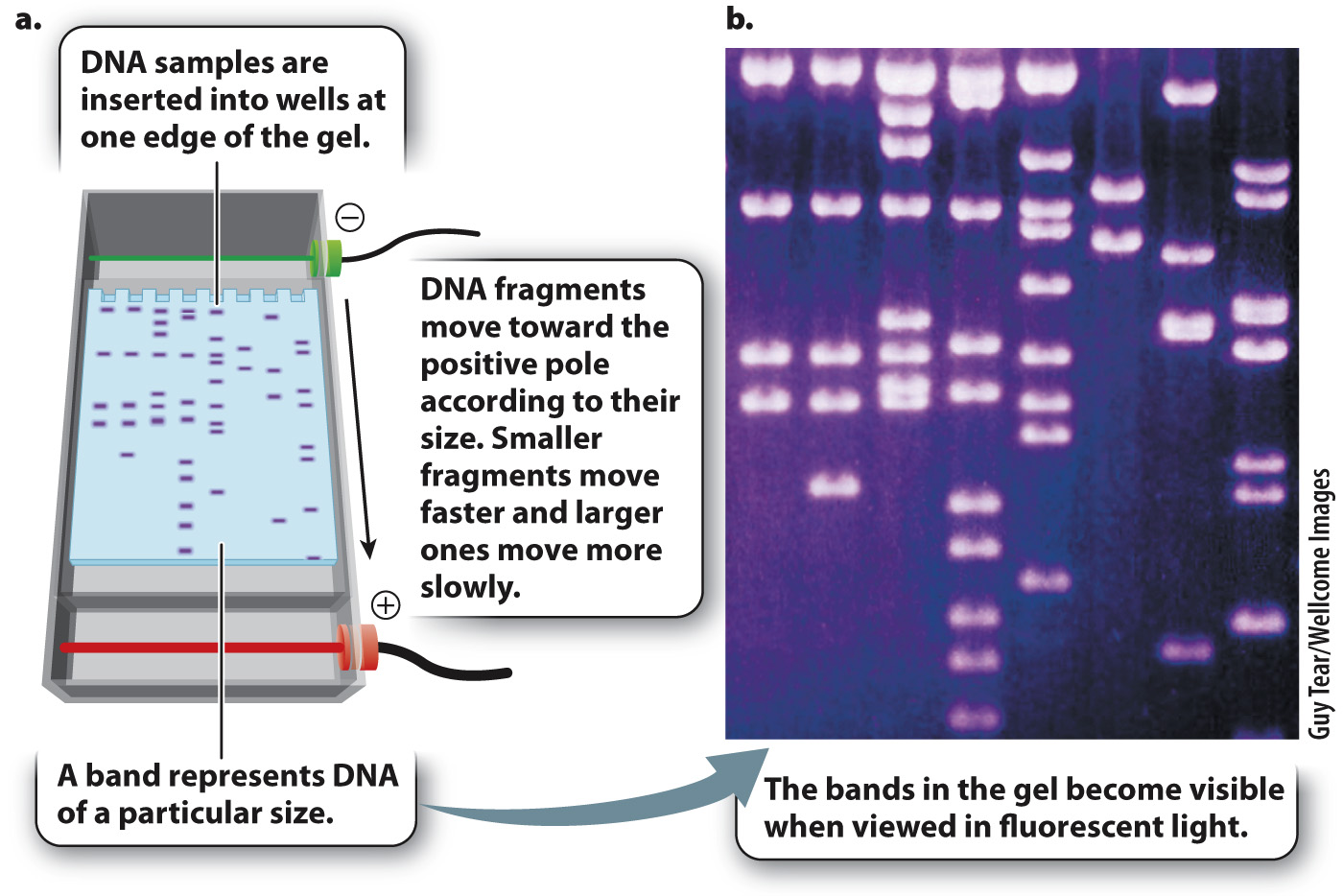
One way to determine the actual size of a DNA fragment is by gel electrophoresis (Fig. 12.14), a procedure in which DNA samples are inserted into slots or wells near the edge of a rectangular slab of porous material resembling solidified agar (the “gel”), which is composed of a tangle of polymers that make it difficult for large molecules to pass through. The gel is then inserted into an apparatus and immersed in a solution that allows an electric current to be passed through it (Fig. 12.14a). Since fragments of double-
260
The DNA molecules move according to their size. Short fragments pass through the pores of the gel more readily than large fragments, and so in a given interval of time short fragments move a greater distance in the gel than large fragments. The rate of migration is dependent only on size, not on sequence, and so all fragments of a given size move together at the same rate in a discrete band, which can be made visible by dyes that bind to DNA and fluoresce under ultraviolet light (Fig. 12.14b). A solution of DNA fragments of known sizes is usually placed in one of the wells, resulting in a series of bands, called a ladder, which can be used for size comparison.
Consider a PCR experiment with 25 base-
Quick Check 2 You do a PCR reaction, run a sample of the product on a gel, and use a dye to visualize the bands. You expect a single product of 300 bp, which you observe, but you also see a band of about 550 bp. Can you suggest a reason why? Does the size of the unexpected band indicate anything about your target sequence?
Quick Check 2 Answer
The most likely reason for observing an unexpected PCR product is that the primers annealed not only to sites flanking the target region but also annealed in the correct orientation to sites flanking a different, nontarget region of the genome. The size of the nontarget region is completely unpredictable, so there is nothing significant in the size of the unexpected product being 550 bp.
Gel electrophoresis can separate DNA fragments produced by any means, not only PCR. Genomic DNA can be cut with certain enzymes and the resulting fragments separated by gel electrophoresis, as described in the next section. Similar procedures can also be used to separate protein molecules.